|
|
@@ -3,21 +3,37 @@ import numpy as np
|
|
|
import matplotlib.pyplot as plt
|
|
|
|
|
|
from filval.result_set import ResultSet
|
|
|
-from filval.histogram import hist, hist2d, hist_add, hist_norm, hist_scale, hist2d_norm
|
|
|
+from filval.histogram import hist, hist2d, hist_add, hist_sub, hist_div, hist_norm, hist_scale, hist2d_norm
|
|
|
from filval.plotting import (decl_plot, render_plots, hist_plot, hist_plot_stack, hist2d_plot,
|
|
|
Plot, generate_dashboard, hists_to_table)
|
|
|
|
|
|
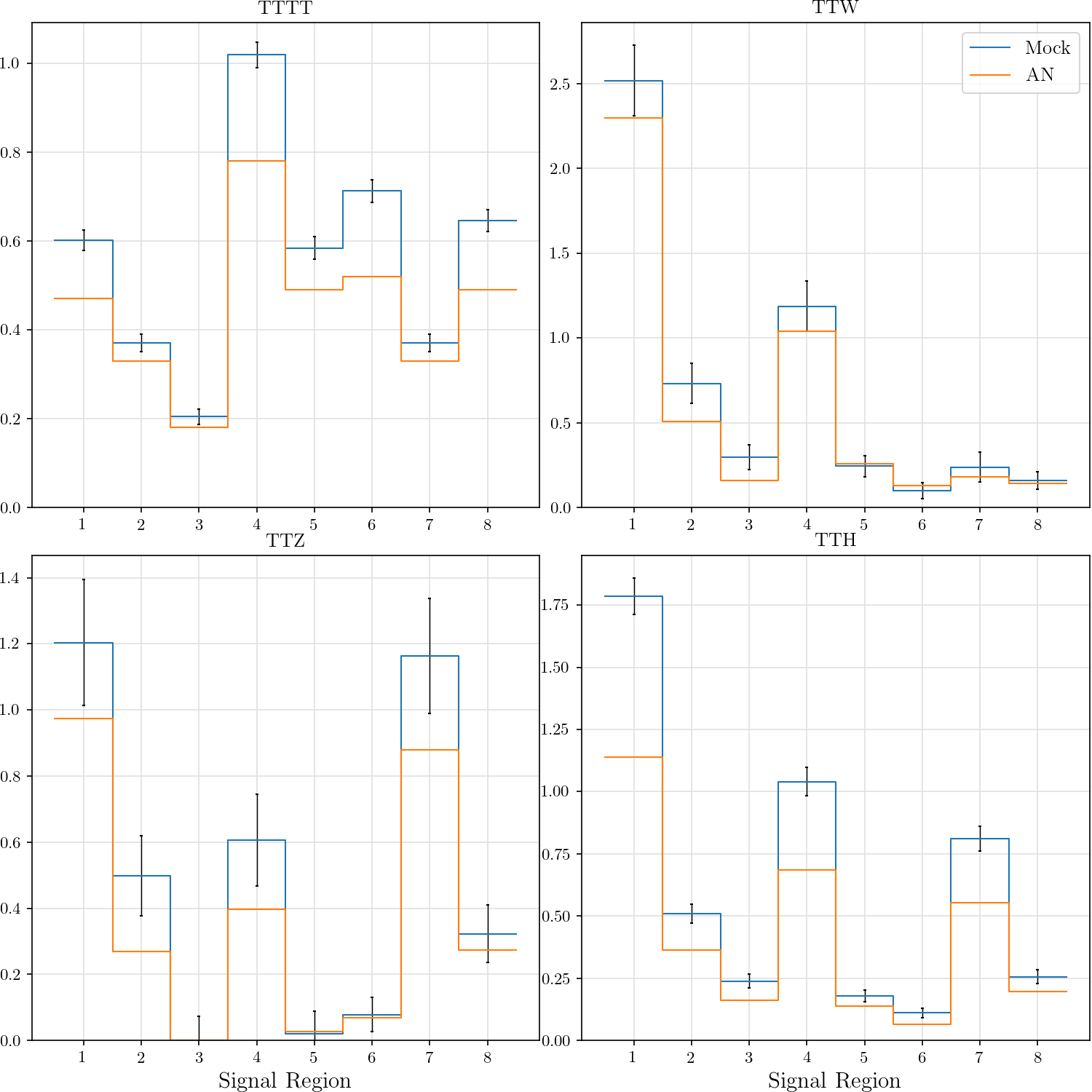
-an_tttt = ([0.47, 0.33, 0.18, 0.78, 0.49, 0.52, 0.33, 0.49],
|
|
|
- [0, 0, 0, 0, 0, 0, 0, 0], [0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5])
|
|
|
-an_ttw = ([2.29663, 0.508494, 0.161166, 1.03811, 0.256401, 0.127582, 0.181522, 0.141659],
|
|
|
- [0, 0, 0, 0, 0, 0, 0, 0], [0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5])
|
|
|
-an_ttz = ([0.974751, 0.269195, 1e-06, 0.395831, 0.0264703, 0.06816, 0.8804, 0.274265],
|
|
|
- [0, 0, 0, 0, 0, 0, 0, 0], [0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5])
|
|
|
-an_tth = ([1.13826, 0.361824, 0.162123, 0.683917, 0.137608, 0.0632719, 0.554491, 0.197864],
|
|
|
- [0, 0, 0, 0, 0, 0, 0, 0], [0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5])
|
|
|
+
|
|
|
+def set_xticks():
|
|
|
+ x_ticks = [f'SR{i}' for i in range(1, 17)]
|
|
|
+ plt.xticks([x for x in range(1, 17)], x_ticks, rotation=60)
|
|
|
+
|
|
|
+
|
|
|
+def get_sr(rs, tau_category, tm=False):
|
|
|
+ if tm:
|
|
|
+ if tau_category == 0:
|
|
|
+ return hist(rs.SRs_0tmtau)
|
|
|
+ if tau_category == 1:
|
|
|
+ return hist(rs.SRs_1tmtau)
|
|
|
+ elif tau_category == 2:
|
|
|
+ return hist(rs.SRs_2tmtau)
|
|
|
+ else:
|
|
|
+ if tau_category == -1:
|
|
|
+ return hist(rs.ignore_tau_SRs)
|
|
|
+ elif tau_category == 0:
|
|
|
+ return hist(rs.SRs_0tau)
|
|
|
+ elif tau_category == 1:
|
|
|
+ return hist(rs.SRs_1tau)
|
|
|
+ elif tau_category == 2:
|
|
|
+ return hist(rs.SRs_2tau)
|
|
|
+
|
|
|
|
|
|
@decl_plot
|
|
|
-def plot_yield_grid(rss, tau_category=-1):
|
|
|
+def plot_yield_grid(tau_category=-1):
|
|
|
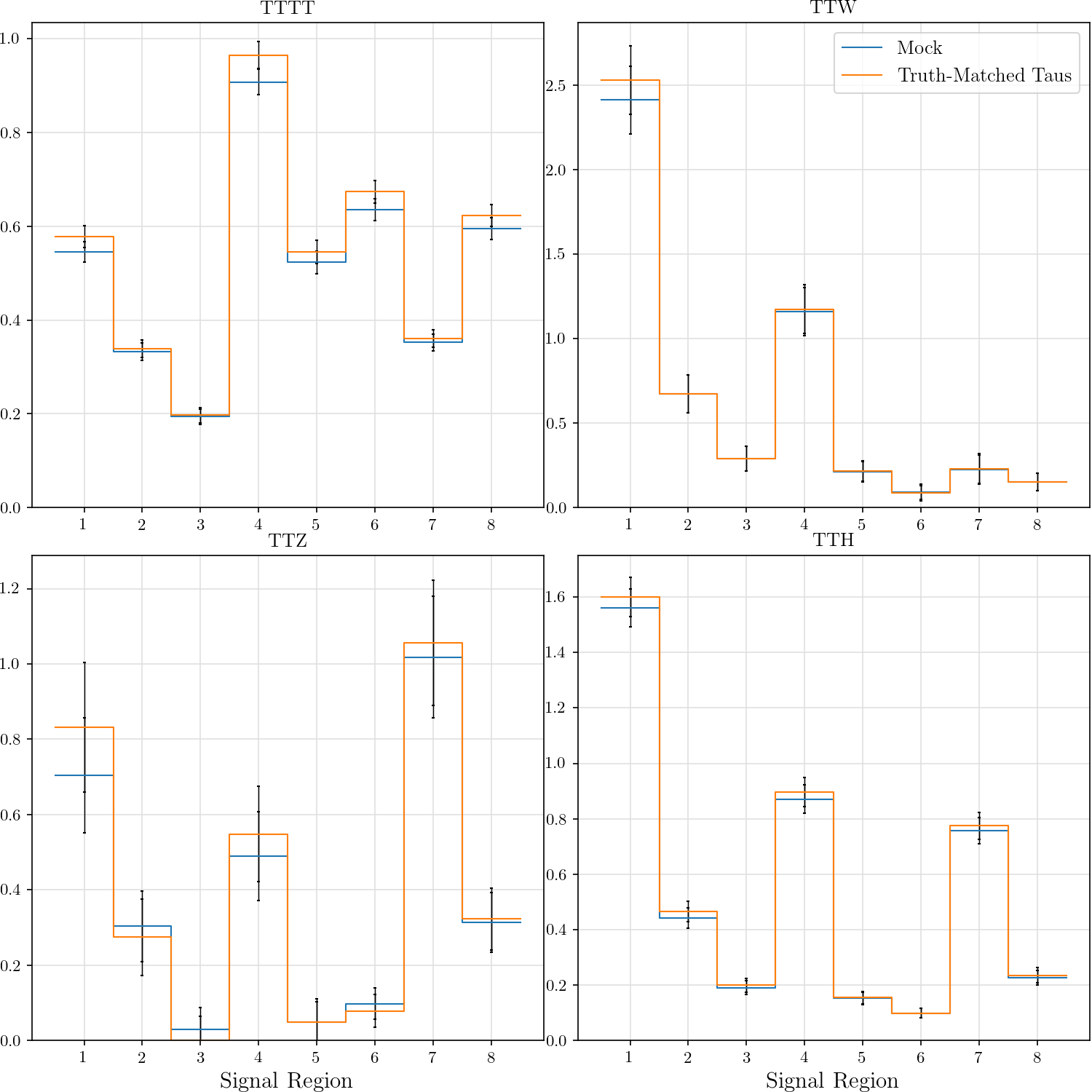
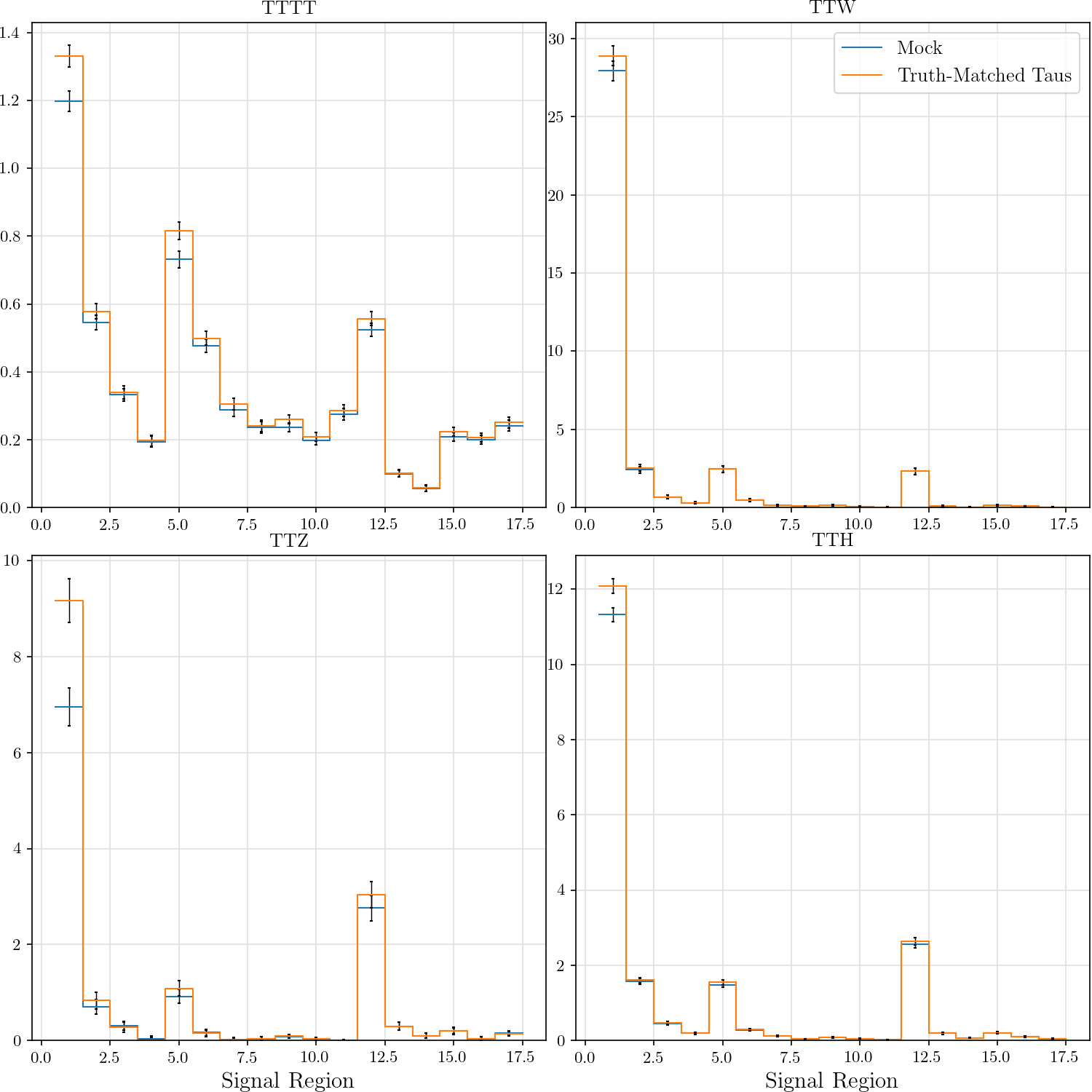
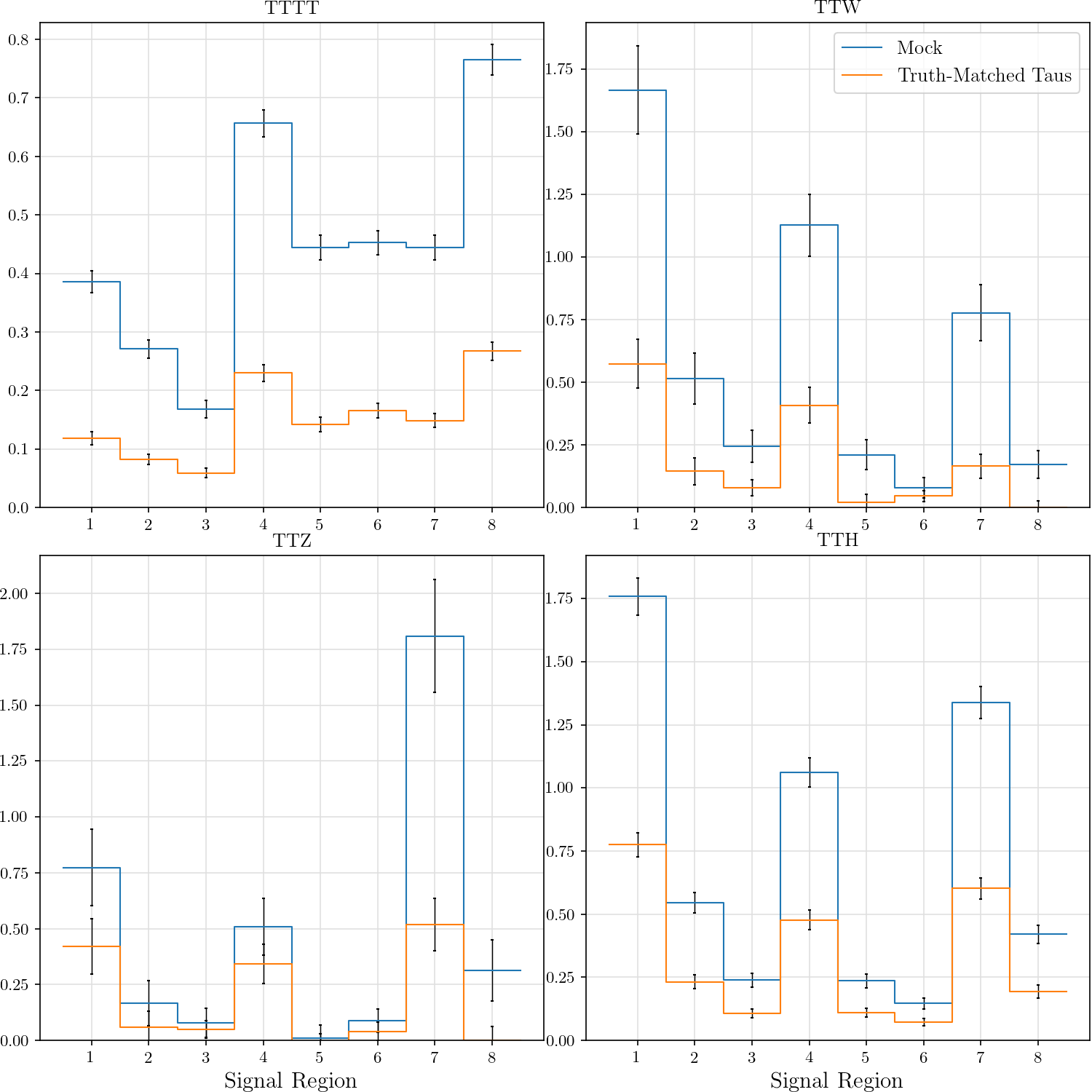
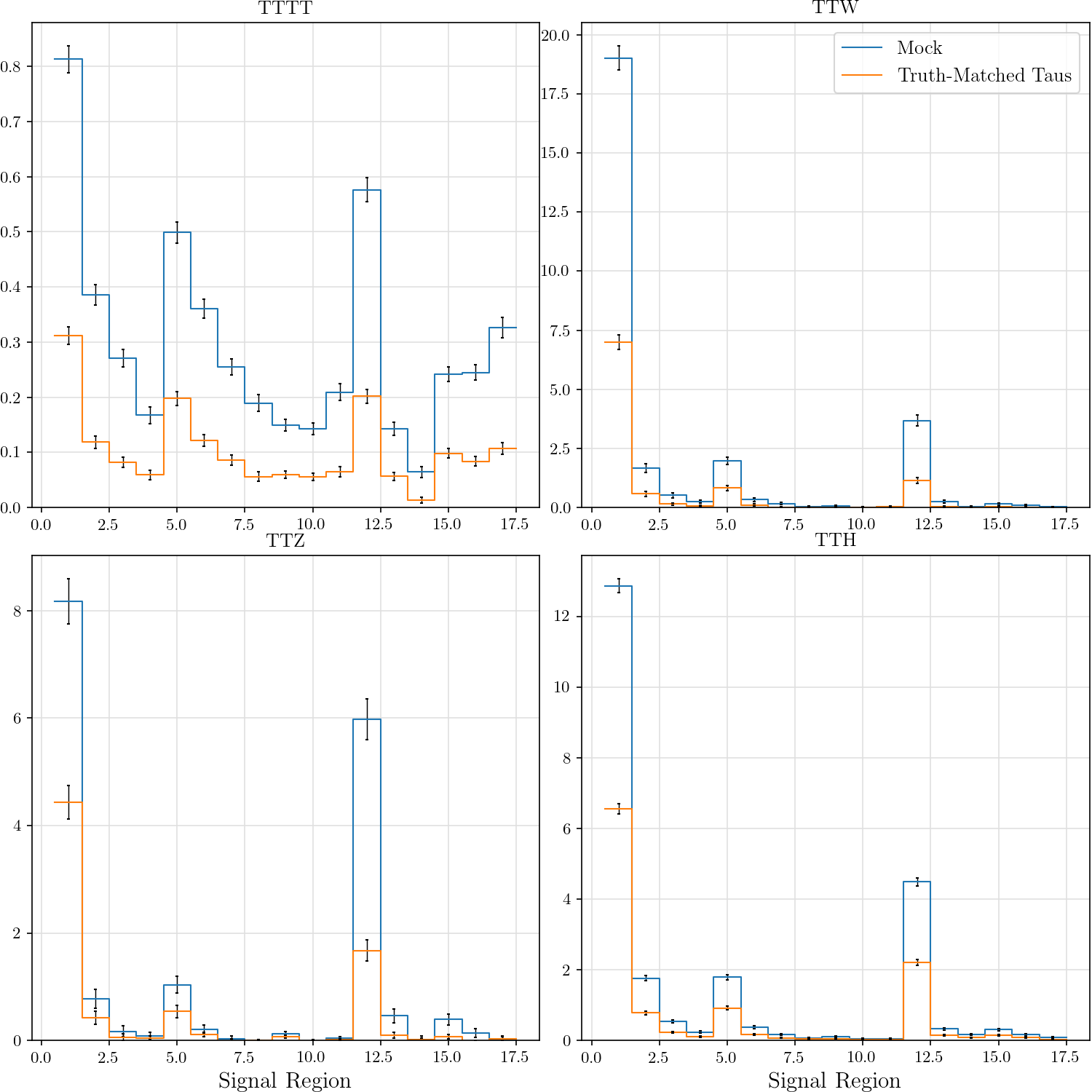
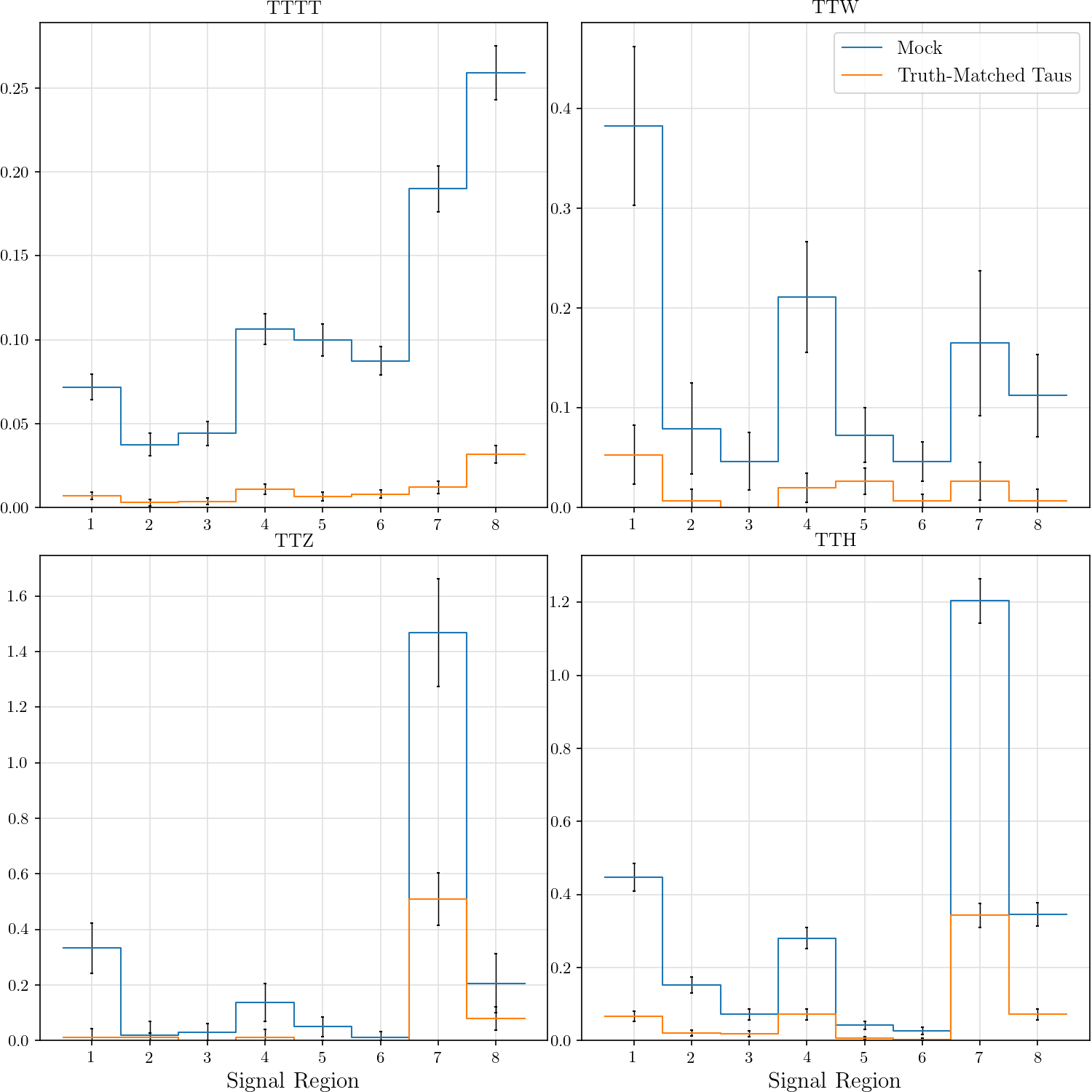
r"""## Event Yield
|
|
|
|
|
|
The event yield for the eight signal regions defined in AN-17-115. Data is normalized
|
|
|
@@ -36,61 +52,43 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
Truth-matched taus are those that match within $\delta R < 0.3$ with gen-level taus that pass the flag `fromHardProcessDecayed`.
|
|
|
"""
|
|
|
|
|
|
- def get_sr(rs, tm=False):
|
|
|
- if tm:
|
|
|
- if tau_category == 0:
|
|
|
- return hist(rs.SRs_0tmtau)
|
|
|
- if tau_category == 1:
|
|
|
- return hist(rs.SRs_1tmtau)
|
|
|
- elif tau_category == 2:
|
|
|
- return hist(rs.SRs_2tmtau)
|
|
|
- else:
|
|
|
- if tau_category == -1:
|
|
|
- return hist(rs.ignore_tau_SRs)
|
|
|
- elif tau_category == 0:
|
|
|
- return hist(rs.SRs_0tau)
|
|
|
- elif tau_category == 1:
|
|
|
- return hist(rs.SRs_1tau)
|
|
|
- elif tau_category == 2:
|
|
|
- return hist(rs.SRs_2tau)
|
|
|
-
|
|
|
_, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
- tttt, ttw, ttz, tth = [get_sr(rs) for rs in rss]
|
|
|
- tm_tttt, tm_ttw, tm_ttz, tm_tth = [get_sr(rs, True) for rs in rss]
|
|
|
+ tttt, ttw, ttz, tth = [get_sr(rs, tau_category) for rs in rss]
|
|
|
+ tm_tttt, tm_ttw, tm_ttz, tm_tth = [get_sr(rs, tau_category, True) for rs in rss]
|
|
|
|
|
|
plt.sca(ax_tttt)
|
|
|
- hist_plot(tttt, title='TTTT', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
- hist_plot(an_tttt, title='TTTT', stats=False, label='AN')
|
|
|
- elif tau_category >= 0:
|
|
|
- hist_plot(tm_tttt, title='TTTT', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
+ plt.title('TTTT')
|
|
|
+ hist_plot(tttt, stats=False, label='Mock Analysis', include_errors=True)
|
|
|
+ if tau_category >= 0:
|
|
|
+ hist_plot(tm_tttt, stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
plt.ylim((0, None))
|
|
|
+ set_xticks()
|
|
|
|
|
|
plt.sca(ax_ttw)
|
|
|
- hist_plot(ttw, title='TTW', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
- hist_plot(an_ttw, title='TTW', stats=False, label='AN')
|
|
|
- elif tau_category >= 0:
|
|
|
- hist_plot(tm_ttw, title='TTW', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
+ plt.title('TTW')
|
|
|
+ hist_plot(ttw, stats=False, label='Mock Analysis', include_errors=True)
|
|
|
+ if tau_category >= 0:
|
|
|
+ hist_plot(tm_ttw, stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
plt.ylim((0, None))
|
|
|
+ set_xticks()
|
|
|
plt.legend()
|
|
|
|
|
|
plt.sca(ax_ttz)
|
|
|
- hist_plot(ttz, title='TTZ', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
- hist_plot(an_ttz, title='TTZ', stats=False, label='AN')
|
|
|
- elif tau_category >= 0:
|
|
|
- hist_plot(tm_ttz, title='TTZ', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
+ plt.title('TTZ')
|
|
|
+ hist_plot(ttz, stats=False, label='Mock Analysis', include_errors=True)
|
|
|
+ if tau_category >= 0:
|
|
|
+ hist_plot(tm_ttz, stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
plt.ylim((0, None))
|
|
|
+ set_xticks()
|
|
|
plt.xlabel('Signal Region')
|
|
|
|
|
|
plt.sca(ax_tth)
|
|
|
- hist_plot(tth, title='TTH', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
- hist_plot(an_tth, title='TTH', stats=False, label='AN')
|
|
|
- elif tau_category >= 0:
|
|
|
- hist_plot(tm_tth, title='TTH', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
+ plt.title('TTH')
|
|
|
+ hist_plot(tth, stats=False, label='Mock Analysis', include_errors=True)
|
|
|
+ if tau_category >= 0:
|
|
|
+ hist_plot(tm_tth, stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
plt.ylim((0, None))
|
|
|
+ set_xticks()
|
|
|
plt.xlabel('Signal Region')
|
|
|
|
|
|
def to_table(hists):
|
|
|
@@ -99,17 +97,14 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
|
|
|
tables = '<h2>Mock</h2>'
|
|
|
tables += to_table([tttt, ttw, ttz, tth])
|
|
|
- if tau_category == -1:
|
|
|
- tables += '<h2>AN</h2>'
|
|
|
- tables += to_table([an_tttt, an_ttw, an_ttz, an_tth])
|
|
|
- elif tau_category >= 0:
|
|
|
+ if tau_category >= 0:
|
|
|
tables += '<h2>TM Taus</h2>'
|
|
|
tables += to_table([tm_tttt, tm_ttw, tm_ttz, tm_tth])
|
|
|
return tables
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
-def plot_yield_v_gen(rss, tau_category=-1):
|
|
|
+def plot_yield_v_gen(tau_category=-1):
|
|
|
r"""## Event Yield Vs. # of Generated Taus
|
|
|
|
|
|
"""
|
|
|
@@ -148,7 +143,7 @@ def plot_yield_v_gen(rss, tau_category=-1):
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
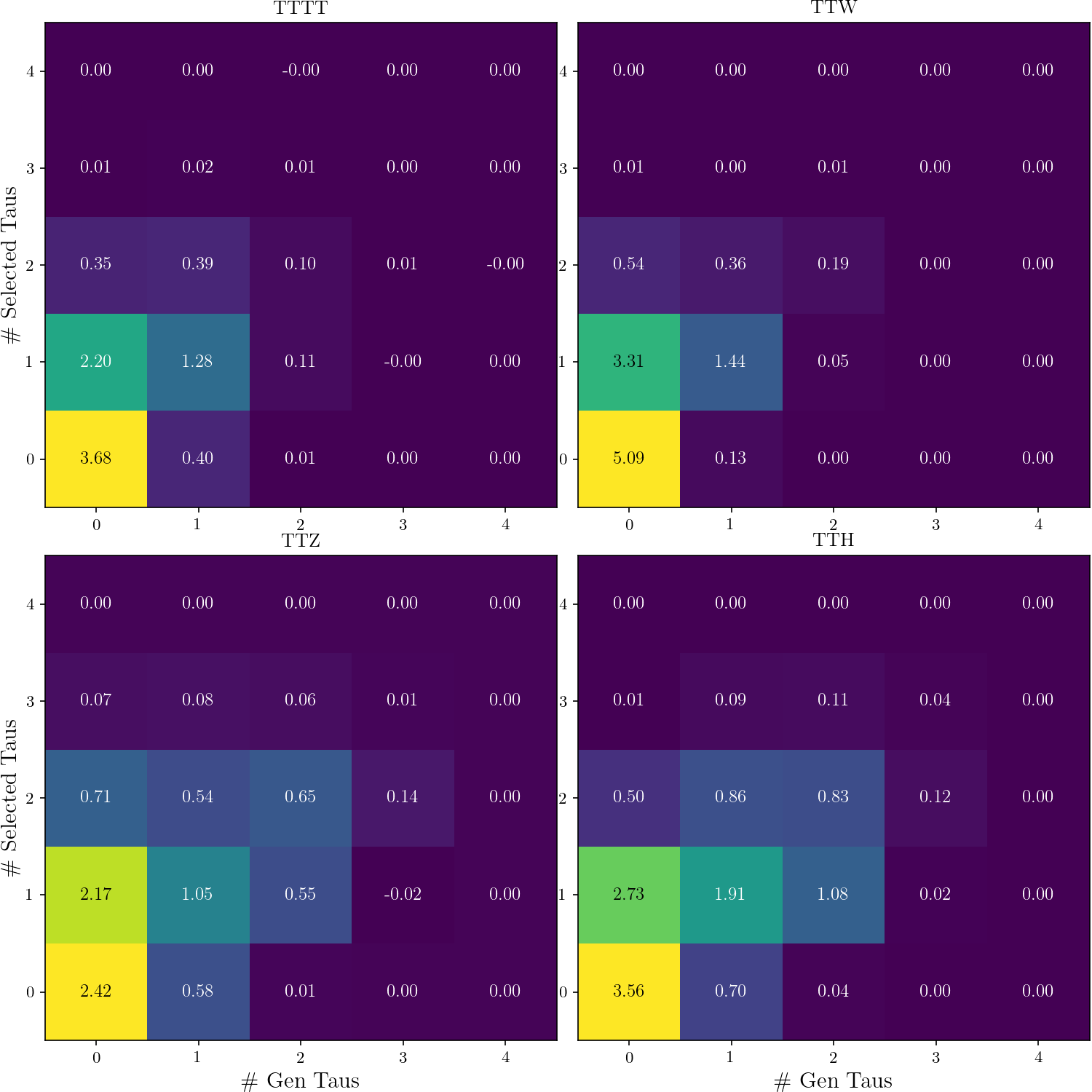
-def plot_nGen_v_nSel(rss):
|
|
|
+def plot_nGen_v_nSel():
|
|
|
_, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
tttt, ttw, ttz, tth = [hist2d(rs.nGen_v_RecoTaus_in_SR) for rs in rss]
|
|
|
|
|
|
@@ -174,7 +169,7 @@ def plot_nGen_v_nSel(rss):
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
-def plot_yield_stack(rss):
|
|
|
+def plot_yield_stack():
|
|
|
r"""## Event Yield - Stacked
|
|
|
|
|
|
The event yield for the eight signal regions defined in AN-17-115. Data is normalized
|
|
|
@@ -192,7 +187,7 @@ def plot_yield_stack(rss):
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
-def plot_lep_multi(rss, dataset):
|
|
|
+def plot_lep_multi(dataset):
|
|
|
_, (ax_els, ax_mus, ax_taus) = plt.subplots(3, 1)
|
|
|
els = list(map(lambda rs: hist_norm(hist(rs.nEls)), rss))
|
|
|
mus = list(map(lambda rs: hist_norm(hist(rs.nMus)), rss))
|
|
|
@@ -217,20 +212,20 @@ def plot_lep_multi(rss, dataset):
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
-def plot_sig_strength(rss):
|
|
|
+def plot_sig_strength(tau_category, tm):
|
|
|
r""" The signal strength of the TTTT signal defined as
|
|
|
|
|
|
- $\frac{S}{\sqrt{S+B}}$
|
|
|
+ $\frac{S}{\sqrt{S+B+\sigma_B^2}}$
|
|
|
|
|
|
"""
|
|
|
- tttt, ttw, ttz, tth = map(lambda rs: hist(rs.SRs), rss)
|
|
|
+ tttt, ttw, ttz, tth = map(lambda rs: get_sr(rs, tau_category, tm), rss)
|
|
|
bg = hist_add(ttw, ttz, tth)
|
|
|
- strength = tttt[0] / np.sqrt(tttt[0] + bg[0])
|
|
|
+ strength = tttt[0] / np.sqrt(tttt[0] + bg[0] + bg[1]**2)
|
|
|
hist_plot((strength, tttt[1], tttt[2]), stats=False)
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
-def plot_event_obs(rss, dataset, in_signal_region=True):
|
|
|
+def plot_event_obs(dataset, in_sr=True):
|
|
|
r"""The distribution of $N_{jet}$, $N_{Bjet}$, MET, and $H_T$ in either all events
|
|
|
or only signal region (igoring taus) events.
|
|
|
|
|
|
@@ -245,7 +240,7 @@ def plot_event_obs(rss, dataset, in_signal_region=True):
|
|
|
|
|
|
def _plot(ax, obs):
|
|
|
plt.sca(ax)
|
|
|
- if in_signal_region:
|
|
|
+ if in_sr:
|
|
|
h = {'MET': rs.met_in_SR,
|
|
|
'HT': rs.ht_in_SR,
|
|
|
'NJET': rs.njet_in_SR,
|
|
|
@@ -255,7 +250,8 @@ def plot_event_obs(rss, dataset, in_signal_region=True):
|
|
|
'HT': rs.ht,
|
|
|
'NJET': rs.njet,
|
|
|
'NBJET': rs.nbjet}[obs]
|
|
|
- hist_plot(hist(h), stats=False, label=dataset, xlabel=obs)
|
|
|
+ hist_plot(hist(h), stats=False, label=dataset)
|
|
|
+ plt.xlabel(obs)
|
|
|
|
|
|
_plot(ax_njet, 'NJET')
|
|
|
_plot(ax_nbjet, 'NBJET')
|
|
|
@@ -264,7 +260,7 @@ def plot_event_obs(rss, dataset, in_signal_region=True):
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
-def plot_event_obs_stack(rss, in_signal_region=True):
|
|
|
+def plot_event_obs_stack(in_signal_region=True):
|
|
|
r"""
|
|
|
|
|
|
"""
|
|
|
@@ -293,25 +289,51 @@ def plot_event_obs_stack(rss, in_signal_region=True):
|
|
|
_plot(ax_ht, 'HT')
|
|
|
_plot(ax_met, 'MET')
|
|
|
|
|
|
-# @decl_plot
|
|
|
-# def plot_s_over_b(rss):
|
|
|
-# def get_sr(rs):
|
|
|
-# h = None
|
|
|
-# if tau_category == 0:
|
|
|
-# h = rs.SRs_0tmtau_diff_nGenTau
|
|
|
-# if tau_category == 1:
|
|
|
-# h = rs.SRs_1tmtau_diff_nGenTau
|
|
|
-# elif tau_category == 2:
|
|
|
-# h = rs.SRs_2tmtau_diff_nGenTau
|
|
|
-# return hist2d_norm(hist2d(h), norm=1, axis=0)
|
|
|
-#
|
|
|
-# _, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
-# tttt =
|
|
|
-# ttw, ttz, tth = [get_sr(rs) for rs in rss]
|
|
|
-# pass
|
|
|
|
|
|
@decl_plot
|
|
|
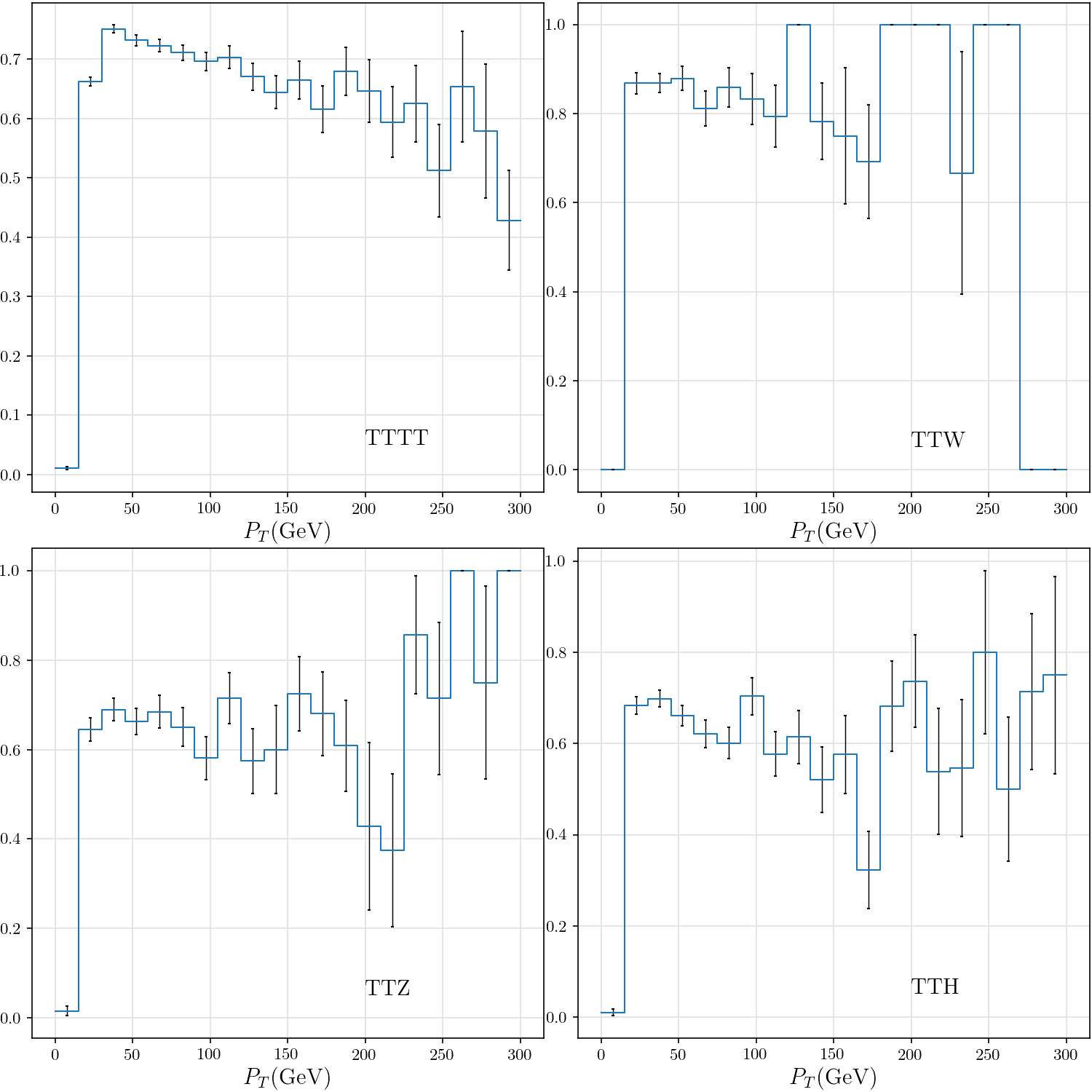
-def plot_tau_efficiency(rss):
|
|
|
+def plot_s_over_b(tau_category):
|
|
|
+
|
|
|
+ tttt, ttw, ttz, tth = [get_sr(rs, tau_category) for rs in rss]
|
|
|
+ sig = get_sr(rss[0], tau_category, tm=True)
|
|
|
+ fake_tttt = hist_sub(tttt, sig)
|
|
|
+
|
|
|
+ total_bg = hist_add(fake_tttt, ttw, ttz, tth)
|
|
|
+
|
|
|
+ plt.subplot(3, 1, 1)
|
|
|
+ hist_plot(total_bg, include_errors=True, label="TTV+TTH+Fake TTTT")
|
|
|
+ hist_plot(sig, include_errors=True, label="Truth Matched TTTT")
|
|
|
+ plt.ylabel('\\# Events')
|
|
|
+ plt.ylim((0, 5.5))
|
|
|
+ set_xticks()
|
|
|
+ plt.legend()
|
|
|
+
|
|
|
+ plt.subplot(3, 1, 2)
|
|
|
+ hist_plot(hist_div(sig, total_bg), label="ratio", color='k')
|
|
|
+ plt.ylabel('Ratio')
|
|
|
+ plt.ylim((0, 2.7))
|
|
|
+ set_xticks()
|
|
|
+
|
|
|
+ plt.subplot(3, 1, 3)
|
|
|
+ bg_vals, bg_errs, _ = total_bg
|
|
|
+ sig_vals, _, _ = sig
|
|
|
+ den = np.sqrt(sig_vals + bg_vals + bg_errs**2)
|
|
|
+ sig = sig_vals / den, *sig[1:]
|
|
|
+
|
|
|
+ hist_plot(sig, color='k')
|
|
|
+ plt.ylabel(r'$S/\sqrt{S+B+\sigma_B^2}$')
|
|
|
+ plt.ylim((0, 0.5))
|
|
|
+ set_xticks()
|
|
|
+
|
|
|
+ # tables = '<h2>Mock</h2>'
|
|
|
+ # tables += to_table([total_bg, ttw, ttz, tth])
|
|
|
+ # if tau_category >= 0:
|
|
|
+ # tables += '<h2>TM Taus</h2>'
|
|
|
+ # tables += to_table([tm_tttt, tm_ttw, tm_ttz, tm_tth])
|
|
|
+ # return tables
|
|
|
+
|
|
|
+
|
|
|
+@decl_plot
|
|
|
+def plot_tau_efficiency():
|
|
|
_, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
tttt, ttw, ttz, tth = list(map(lambda rs: hist(rs.tau_efficiency_v_pt), rss))
|
|
|
|
|
|
@@ -324,14 +346,16 @@ def plot_tau_efficiency(rss):
|
|
|
hist_plot(h, stats=False, label=dataset, include_errors=True)
|
|
|
plt.text(200, 0.05, dataset)
|
|
|
plt.xlabel(r"$P_T$(GeV)")
|
|
|
+ plt.ylim((0, 1.1))
|
|
|
|
|
|
_plot(ax_tttt, 'TTTT')
|
|
|
_plot(ax_ttw, 'TTW')
|
|
|
_plot(ax_ttz, 'TTZ')
|
|
|
_plot(ax_tth, 'TTH')
|
|
|
|
|
|
+
|
|
|
@decl_plot
|
|
|
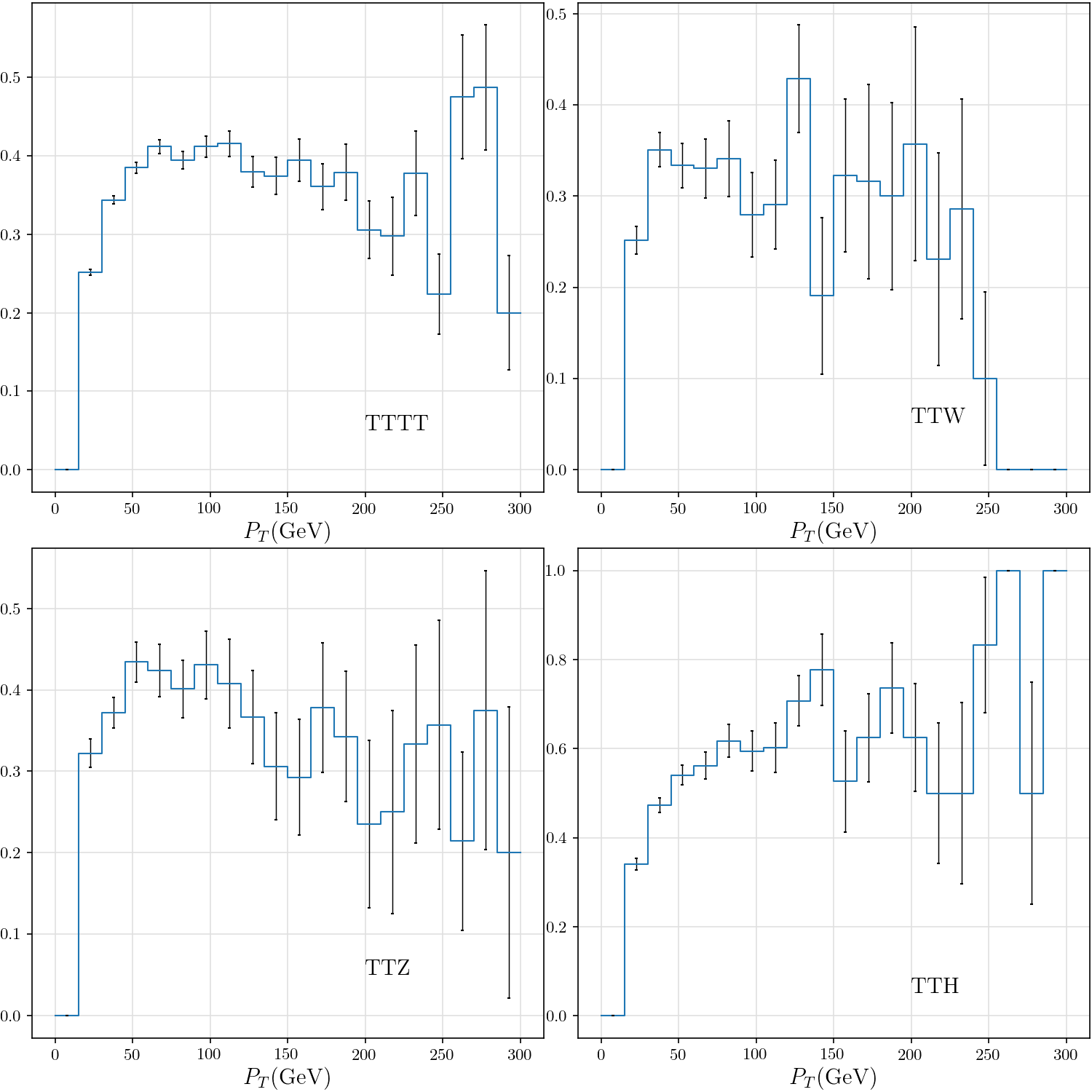
-def plot_tau_purity(rss):
|
|
|
+def plot_tau_purity():
|
|
|
_, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
tttt, ttw, ttz, tth = list(map(lambda rs: hist(rs.tau_purity_v_pt), rss))
|
|
|
|
|
|
@@ -344,6 +368,7 @@ def plot_tau_purity(rss):
|
|
|
hist_plot(h, stats=False, label=dataset, include_errors=True)
|
|
|
plt.text(200, 0.05, dataset)
|
|
|
plt.xlabel(r"$P_T$(GeV)")
|
|
|
+ plt.ylim((0, 1.1))
|
|
|
|
|
|
_plot(ax_tttt, 'TTTT')
|
|
|
_plot(ax_ttw, 'TTW')
|
|
|
@@ -352,119 +377,80 @@ def plot_tau_purity(rss):
|
|
|
|
|
|
|
|
|
if __name__ == '__main__':
|
|
|
- data_path = 'data/output_testing/'
|
|
|
+ ext = ""
|
|
|
+ # ext = "_dr03"
|
|
|
+ data_path = f'data/TauStudies/output{ext}/'
|
|
|
save_plots = False
|
|
|
- # First create a ResultSet object which loads all of the objects from root file
|
|
|
- # into memory and makes them available as attributes
|
|
|
+
|
|
|
rss = (ResultSet("tttt", data_path+'yield_tttt.root'),
|
|
|
ResultSet("ttw", data_path+'yield_ttw.root'),
|
|
|
ResultSet("ttz", data_path+'yield_ttz.root'),
|
|
|
ResultSet("tth", data_path+'yield_tth.root'))
|
|
|
|
|
|
- # Next, declare all of the (sub)plots that will be assembled into full
|
|
|
- # figures later
|
|
|
- yield_tau_ignore_tau = plot_yield_grid, (rss, -1)
|
|
|
- yield_tau_0tau = plot_yield_grid, (rss, 0)
|
|
|
- yield_tau_1tau = plot_yield_grid, (rss, 1)
|
|
|
- yield_tau_2tau = plot_yield_grid, (rss, 2)
|
|
|
-
|
|
|
- tttt_event_obs_in_sr = plot_event_obs, (rss, 'TTTT'), {'in_signal_region': True}
|
|
|
- ttw_event_obs_in_sr = plot_event_obs, (rss, 'TTW'), {'in_signal_region': True}
|
|
|
- ttz_event_obs_in_sr = plot_event_obs, (rss, 'TTZ'), {'in_signal_region': True}
|
|
|
- tth_event_obs_in_sr = plot_event_obs, (rss, 'TTH'), {'in_signal_region': True}
|
|
|
+ tttt_event_obs_in_sr = plot_event_obs, ('TTTT',), {'in_sr': True}
|
|
|
+ ttw_event_obs_in_sr = plot_event_obs, ('TTW',), {'in_sr': True}
|
|
|
+ ttz_event_obs_in_sr = plot_event_obs, ('TTZ',), {'in_sr': True}
|
|
|
+ tth_event_obs_in_sr = plot_event_obs, ('TTH',), {'in_sr': True}
|
|
|
|
|
|
- tttt_event_obs = plot_event_obs, (rss, 'TTTT'), {'in_signal_region': False}
|
|
|
- ttw_event_obs = plot_event_obs, (rss, 'TTW'), {'in_signal_region': False}
|
|
|
- ttz_event_obs = plot_event_obs, (rss, 'TTZ'), {'in_signal_region': False}
|
|
|
- tth_event_obs = plot_event_obs, (rss, 'TTH'), {'in_signal_region': False}
|
|
|
+ tttt_event_obs = plot_event_obs, ('TTTT',), {'in_sr': False}
|
|
|
+ ttw_event_obs = plot_event_obs, ('TTW',), {'in_sr': False}
|
|
|
+ ttz_event_obs = plot_event_obs, ('TTZ',), {'in_sr': False}
|
|
|
+ tth_event_obs = plot_event_obs, ('TTH',), {'in_sr': False}
|
|
|
|
|
|
- tttt_lep_multi = plot_lep_multi, (rss, 'TTTT')
|
|
|
- ttw_lep_multi = plot_lep_multi, (rss, 'TTW')
|
|
|
- ttz_lep_multi = plot_lep_multi, (rss, 'TTZ')
|
|
|
- tth_lep_multi = plot_lep_multi, (rss, 'TTH')
|
|
|
+ tttt_lep_multi = plot_lep_multi, ('TTTT',)
|
|
|
+ ttw_lep_multi = plot_lep_multi, ('TTW',)
|
|
|
+ ttz_lep_multi = plot_lep_multi, ('TTZ',)
|
|
|
+ tth_lep_multi = plot_lep_multi, ('TTH',)
|
|
|
|
|
|
- yield_v_gen_0tm = plot_yield_v_gen, (rss, 0)
|
|
|
- yield_v_gen_1tm = plot_yield_v_gen, (rss, 1)
|
|
|
- yield_v_gen_2tm = plot_yield_v_gen, (rss, 2)
|
|
|
- nGen_v_nSel = plot_nGen_v_nSel, (rss,)
|
|
|
-
|
|
|
- tau_efficiency = plot_tau_efficiency, (rss, )
|
|
|
- tau_purity = plot_tau_purity, (rss, )
|
|
|
+ yield_v_gen_0tm = plot_yield_v_gen, (0,)
|
|
|
+ yield_v_gen_1tm = plot_yield_v_gen, (1,)
|
|
|
+ yield_v_gen_2tm = plot_yield_v_gen, (2,)
|
|
|
+ nGen_v_nSel = plot_nGen_v_nSel
|
|
|
|
|
|
# Now assemble the plots into figures.
|
|
|
plots = [
|
|
|
- Plot([[yield_tau_ignore_tau]],
|
|
|
+
|
|
|
+ Plot((plot_yield_grid, (-1,)),
|
|
|
'Yield Ignoring Taus'),
|
|
|
- Plot([[yield_tau_0tau]],
|
|
|
- 'Yield For events with 0 Tau'),
|
|
|
- Plot([[yield_tau_1tau]],
|
|
|
- 'Yield For events with 1 Tau'),
|
|
|
- Plot([[yield_tau_2tau]],
|
|
|
- 'Yield For events with 2 or more Tau'),
|
|
|
-
|
|
|
- # Plot([[yield_v_gen_0tm]],
|
|
|
- # 'Yield For events with 0 TM Tau Vs Gen Tau'),
|
|
|
- # Plot([[yield_v_gen_1tm]],
|
|
|
- # 'Yield For events with 1 TM Tau Vs Gen Tau'),
|
|
|
- # Plot([[yield_v_gen_2tm]],
|
|
|
- # 'Yield For events with 2 TM Tau Vs Gen Tau'),
|
|
|
- Plot([[nGen_v_nSel]],
|
|
|
+ Plot((plot_yield_grid, (0,)), 'Yield For events with 0 Tau'),
|
|
|
+ Plot((plot_s_over_b, (0,)), 'Real TTTT and Total BG - 0 Tau'),
|
|
|
+ Plot((plot_yield_grid, (1,)), 'Yield For events with 1 Tau'),
|
|
|
+ Plot((plot_s_over_b, (1,)), 'Real TTTT and Total BG - 1 Tau'),
|
|
|
+ Plot((plot_yield_grid, (2,)), 'Yield For events with 2 or more Tau'),
|
|
|
+ Plot((plot_s_over_b, (2,)), 'Real TTTT and Total BG - 2 or more Tau'),
|
|
|
+ Plot(nGen_v_nSel,
|
|
|
r'#Generated tau vs. #Selected tau'),
|
|
|
-
|
|
|
- Plot([[tttt_lep_multi]],
|
|
|
+ Plot(tttt_lep_multi,
|
|
|
'Lepton Multiplicity - TTTT'),
|
|
|
- Plot([[ttw_lep_multi]],
|
|
|
+ Plot(ttw_lep_multi,
|
|
|
'Lepton Multiplicity - TTW'),
|
|
|
- Plot([[ttz_lep_multi]],
|
|
|
+ Plot(ttz_lep_multi,
|
|
|
'Lepton Multiplicity - TTZ'),
|
|
|
- Plot([[tth_lep_multi]],
|
|
|
+ Plot(tth_lep_multi,
|
|
|
'Lepton Multiplicity - TTH'),
|
|
|
- Plot([[tttt_event_obs_in_sr]],
|
|
|
+ Plot(tttt_event_obs_in_sr,
|
|
|
'TTTT - Event Observables (In SR)'),
|
|
|
- Plot([[ttw_event_obs_in_sr]],
|
|
|
+ Plot(ttw_event_obs_in_sr,
|
|
|
'TTW - Event Observables (In SR)'),
|
|
|
- Plot([[ttz_event_obs_in_sr]],
|
|
|
+ Plot(ttz_event_obs_in_sr,
|
|
|
'TTZ - Event Observables (In SR)'),
|
|
|
- Plot([[tth_event_obs_in_sr]],
|
|
|
+ Plot(tth_event_obs_in_sr,
|
|
|
'TTH - Event Observables (In SR)'),
|
|
|
|
|
|
- Plot([[tau_efficiency]],
|
|
|
+ Plot(plot_tau_efficiency,
|
|
|
'Tau Efficiency'),
|
|
|
- Plot([[tau_purity]],
|
|
|
+ Plot(plot_tau_purity,
|
|
|
'Tau Purity'),
|
|
|
- # Plot([[tttt_event_obs]],
|
|
|
- # 'TTTT - Event Observables (All Events)'),
|
|
|
- # Plot([[ttw_event_obs]],
|
|
|
- # 'TTW - Event Observables (All Events)'),
|
|
|
- # Plot([[ttz_event_obs]],
|
|
|
- # 'TTZ - Event Observables (All Events)'),
|
|
|
- # Plot([[tth_event_obs]],
|
|
|
- # 'TTH - Event Observables (All Events)'),
|
|
|
-
|
|
|
- # Plot([[yield_notau]],
|
|
|
- # 'Yield Without Tau'),
|
|
|
- # Plot([[yield_tau_stack]],
|
|
|
- # 'Yield With Tau Stacked'),
|
|
|
- # Plot([[yield_notau_stack]],
|
|
|
- # 'Yield Without Tau Stacked'),
|
|
|
- # Plot([[yield_tau_stack],
|
|
|
- # [yield_notau_stack]],
|
|
|
- # 'Event Yield, top: with tau, bottom: no tau'),
|
|
|
- # Plot([[sig_strength_tau],
|
|
|
- # [sig_strength_notau]],
|
|
|
- # 'Signal Strength'),
|
|
|
- # Plot([[event_obs_stack]],
|
|
|
- # 'Event Observables'),
|
|
|
]
|
|
|
|
|
|
# Finally, render and save the plots and generate the html+bootstrap
|
|
|
# dashboard to view them
|
|
|
render_plots(plots, to_disk=save_plots)
|
|
|
if not save_plots:
|
|
|
- generate_dashboard(plots, 'TTTT Yields',
|
|
|
- output='yields.html',
|
|
|
+ with open(data_path+'config.yaml') as f:
|
|
|
+ config_txt = f.read()
|
|
|
+ generate_dashboard(plots, 'Tau Studies',
|
|
|
+ output=f'tau_studies{ext}.html',
|
|
|
source=__file__,
|
|
|
- ana_source=("https://github.com/cfangmeier/FTAnalysis/commit/"
|
|
|
- "0cbdac4509391fffb9fff87d0521b7dd0a30a55c"),
|
|
|
- config=data_path+'config.yaml'
|
|
|
+ config=config_txt
|
|
|
)
|