|
|
@@ -310,6 +310,26 @@ def plot_event_obs_stack(rss, in_signal_region=True):
|
|
|
# ttw, ttz, tth = [get_sr(rs) for rs in rss]
|
|
|
# pass
|
|
|
|
|
|
+@decl_plot
|
|
|
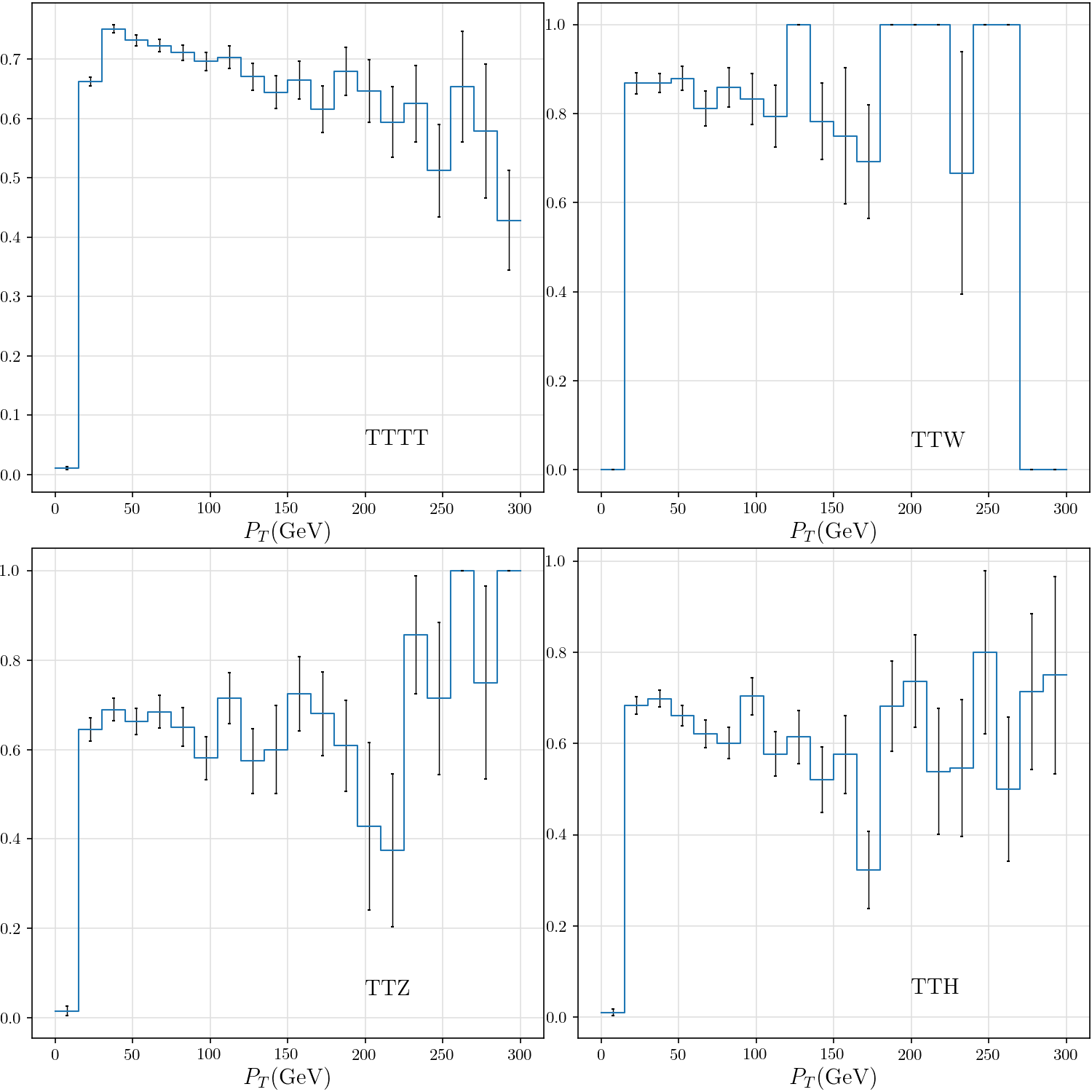
+def plot_tau_efficiency(rss):
|
|
|
+ _, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
+ tttt, ttw, ttz, tth = list(map(lambda rs: hist(rs.tau_efficiency_v_pt), rss))
|
|
|
+
|
|
|
+ def _plot(ax, dataset):
|
|
|
+ plt.sca(ax)
|
|
|
+ h = {'TTTT': tttt,
|
|
|
+ 'TTW': ttw,
|
|
|
+ 'TTZ': ttz,
|
|
|
+ 'TTH': tth}[dataset]
|
|
|
+ hist_plot(h, stats=False, label=dataset, include_errors=True)
|
|
|
+ plt.text(200, 0.05, dataset)
|
|
|
+ plt.xlabel(r"$P_T$(GeV)")
|
|
|
+
|
|
|
+ _plot(ax_tttt, 'TTTT')
|
|
|
+ _plot(ax_ttw, 'TTW')
|
|
|
+ _plot(ax_ttz, 'TTZ')
|
|
|
+ _plot(ax_tth, 'TTH')
|
|
|
+
|
|
|
@decl_plot
|
|
|
def plot_tau_purity(rss):
|
|
|
_, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
@@ -321,7 +341,7 @@ def plot_tau_purity(rss):
|
|
|
'TTW': ttw,
|
|
|
'TTZ': ttz,
|
|
|
'TTH': tth}[dataset]
|
|
|
- hist_plot(h, stats=False, label=dataset)
|
|
|
+ hist_plot(h, stats=False, label=dataset, include_errors=True)
|
|
|
plt.text(200, 0.05, dataset)
|
|
|
plt.xlabel(r"$P_T$(GeV)")
|
|
|
|
|
|
@@ -332,8 +352,8 @@ def plot_tau_purity(rss):
|
|
|
|
|
|
|
|
|
if __name__ == '__main__':
|
|
|
- data_path = 'data/output_new_sr_new_id_binning/'
|
|
|
- save_plots = True
|
|
|
+ data_path = 'data/output_testing/'
|
|
|
+ save_plots = False
|
|
|
# First create a ResultSet object which loads all of the objects from root file
|
|
|
# into memory and makes them available as attributes
|
|
|
rss = (ResultSet("tttt", data_path+'yield_tttt.root'),
|
|
|
@@ -366,9 +386,11 @@ if __name__ == '__main__':
|
|
|
yield_v_gen_0tm = plot_yield_v_gen, (rss, 0)
|
|
|
yield_v_gen_1tm = plot_yield_v_gen, (rss, 1)
|
|
|
yield_v_gen_2tm = plot_yield_v_gen, (rss, 2)
|
|
|
- # tau_purity = plot_tau_purity, (rss)
|
|
|
nGen_v_nSel = plot_nGen_v_nSel, (rss,)
|
|
|
|
|
|
+ tau_efficiency = plot_tau_efficiency, (rss, )
|
|
|
+ tau_purity = plot_tau_purity, (rss, )
|
|
|
+
|
|
|
# Now assemble the plots into figures.
|
|
|
plots = [
|
|
|
Plot([[yield_tau_ignore_tau]],
|
|
|
@@ -405,6 +427,11 @@ if __name__ == '__main__':
|
|
|
'TTZ - Event Observables (In SR)'),
|
|
|
Plot([[tth_event_obs_in_sr]],
|
|
|
'TTH - Event Observables (In SR)'),
|
|
|
+
|
|
|
+ Plot([[tau_efficiency]],
|
|
|
+ 'Tau Efficiency'),
|
|
|
+ Plot([[tau_purity]],
|
|
|
+ 'Tau Purity'),
|
|
|
# Plot([[tttt_event_obs]],
|
|
|
# 'TTTT - Event Observables (All Events)'),
|
|
|
# Plot([[ttw_event_obs]],
|
|
|
@@ -428,8 +455,6 @@ if __name__ == '__main__':
|
|
|
# 'Signal Strength'),
|
|
|
# Plot([[event_obs_stack]],
|
|
|
# 'Event Observables'),
|
|
|
- # Plot([[tau_purity]],
|
|
|
- # 'Tau Purity'),
|
|
|
]
|
|
|
|
|
|
# Finally, render and save the plots and generate the html+bootstrap
|