|
|
@@ -3,9 +3,9 @@ import numpy as np
|
|
|
import matplotlib.pyplot as plt
|
|
|
|
|
|
from filval.result_set import ResultSet
|
|
|
-from filval.histogram_utils import hist, hist_add, hist_normalize, hist_scale
|
|
|
-from filval.plotter import (decl_plot, render_plots, hist_plot, hist_plot_stack,
|
|
|
- Plot, generate_dashboard, hists_to_table)
|
|
|
+from filval.histogram import hist, hist2d, hist_add, hist_norm, hist_scale, hist2d_norm
|
|
|
+from filval.plotting import (decl_plot, render_plots, hist_plot, hist_plot_stack, hist2d_plot,
|
|
|
+ Plot, generate_dashboard, hists_to_table)
|
|
|
|
|
|
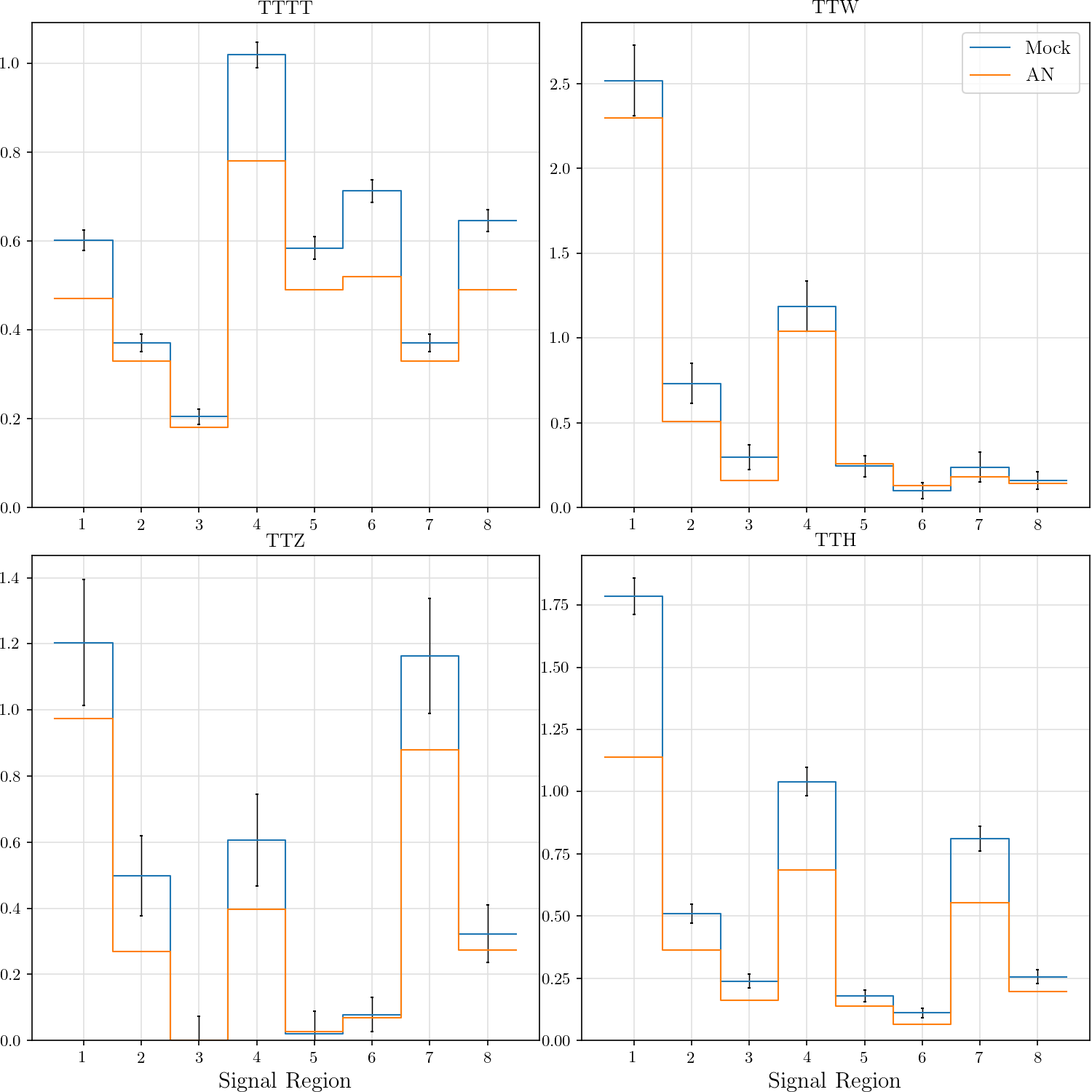
an_tttt = ([0.47, 0.33, 0.18, 0.78, 0.49, 0.52, 0.33, 0.49],
|
|
|
[0, 0, 0, 0, 0, 0, 0, 0], [0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5])
|
|
|
@@ -60,7 +60,7 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
|
|
|
plt.sca(ax_tttt)
|
|
|
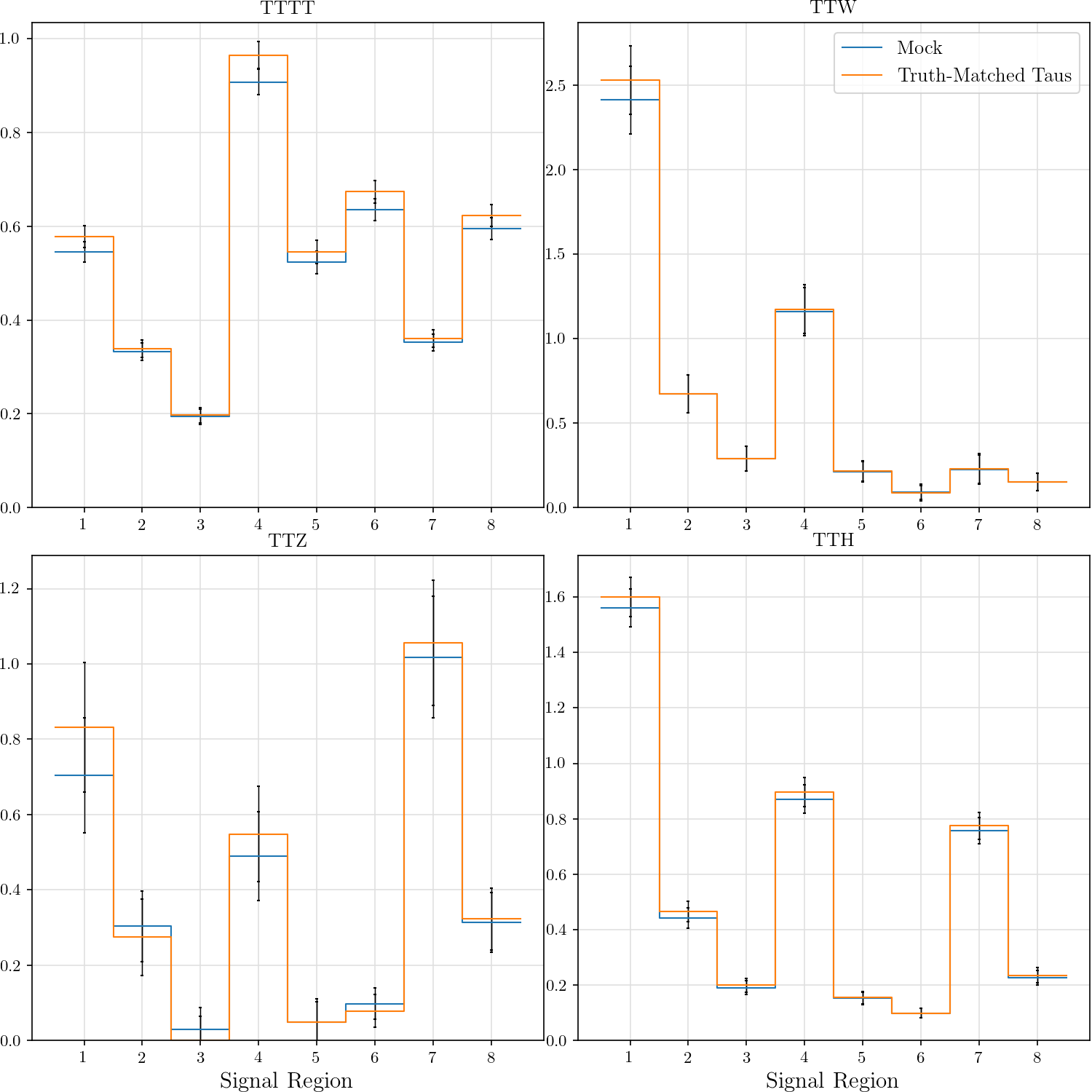
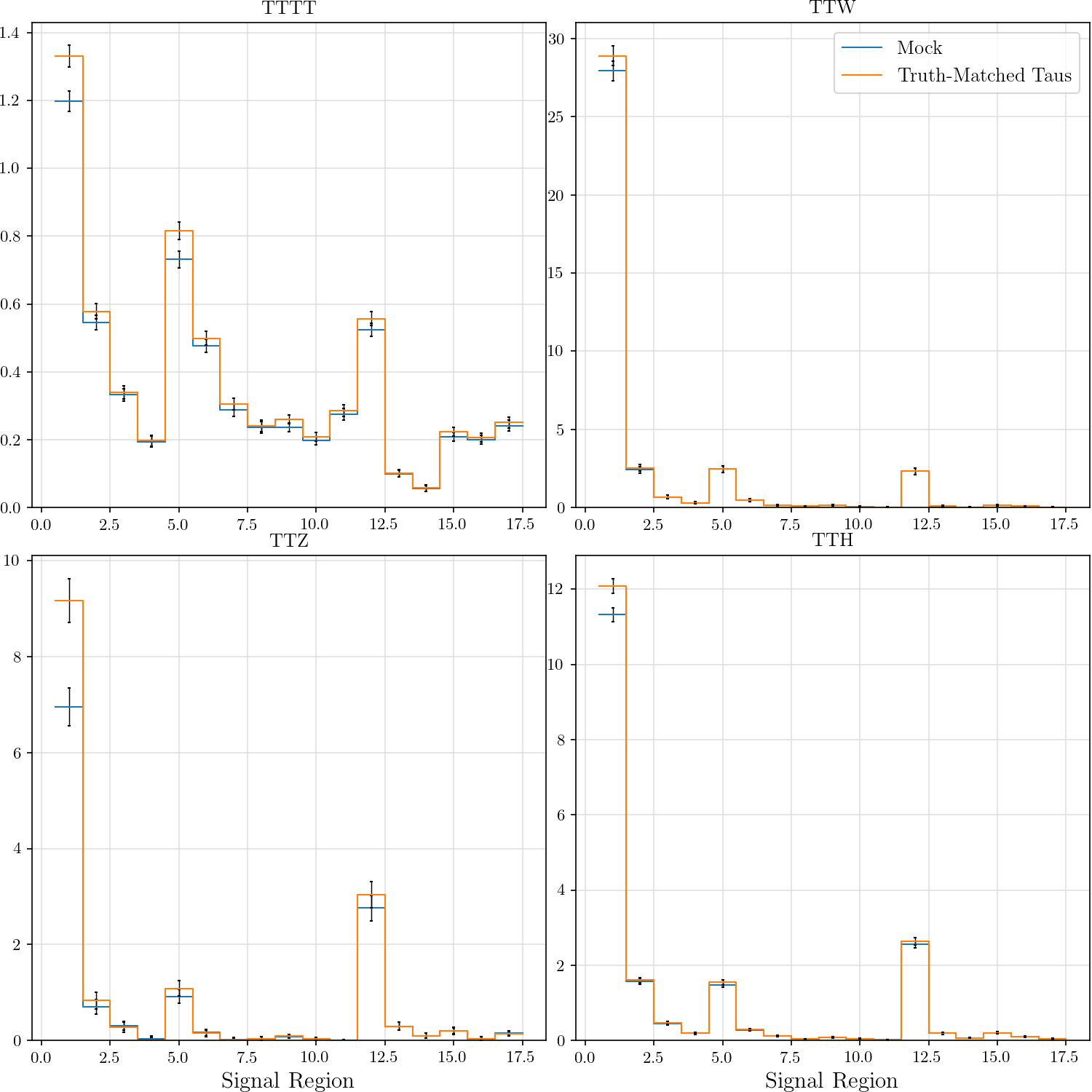
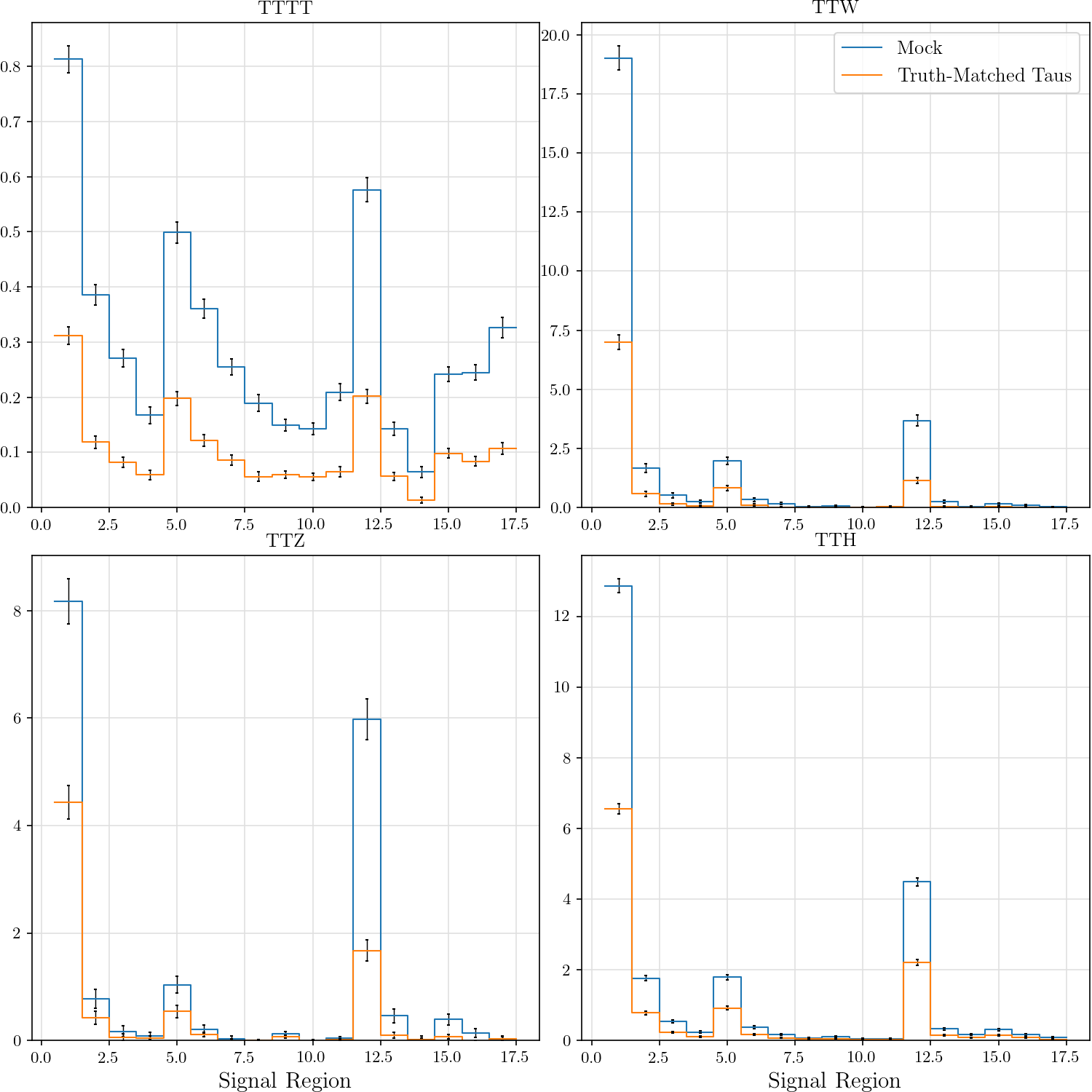
hist_plot(tttt, title='TTTT', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1:
|
|
|
+ if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
hist_plot(an_tttt, title='TTTT', stats=False, label='AN')
|
|
|
elif tau_category >= 0:
|
|
|
hist_plot(tm_tttt, title='TTTT', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
@@ -68,7 +68,7 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
|
|
|
plt.sca(ax_ttw)
|
|
|
hist_plot(ttw, title='TTW', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1:
|
|
|
+ if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
hist_plot(an_ttw, title='TTW', stats=False, label='AN')
|
|
|
elif tau_category >= 0:
|
|
|
hist_plot(tm_ttw, title='TTW', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
@@ -77,7 +77,7 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
|
|
|
plt.sca(ax_ttz)
|
|
|
hist_plot(ttz, title='TTZ', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1:
|
|
|
+ if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
hist_plot(an_ttz, title='TTZ', stats=False, label='AN')
|
|
|
elif tau_category >= 0:
|
|
|
hist_plot(tm_ttz, title='TTZ', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
@@ -86,7 +86,7 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
|
|
|
plt.sca(ax_tth)
|
|
|
hist_plot(tth, title='TTH', stats=False, label='Mock', include_errors=True)
|
|
|
- if tau_category == -1:
|
|
|
+ if tau_category == -1 and len(tttt[0]) == len(an_tttt[0]):
|
|
|
hist_plot(an_tth, title='TTH', stats=False, label='AN')
|
|
|
elif tau_category >= 0:
|
|
|
hist_plot(tm_tth, title='TTH', stats=False, label='Truth-Matched Taus', include_errors=True)
|
|
|
@@ -95,7 +95,7 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
|
|
|
def to_table(hists):
|
|
|
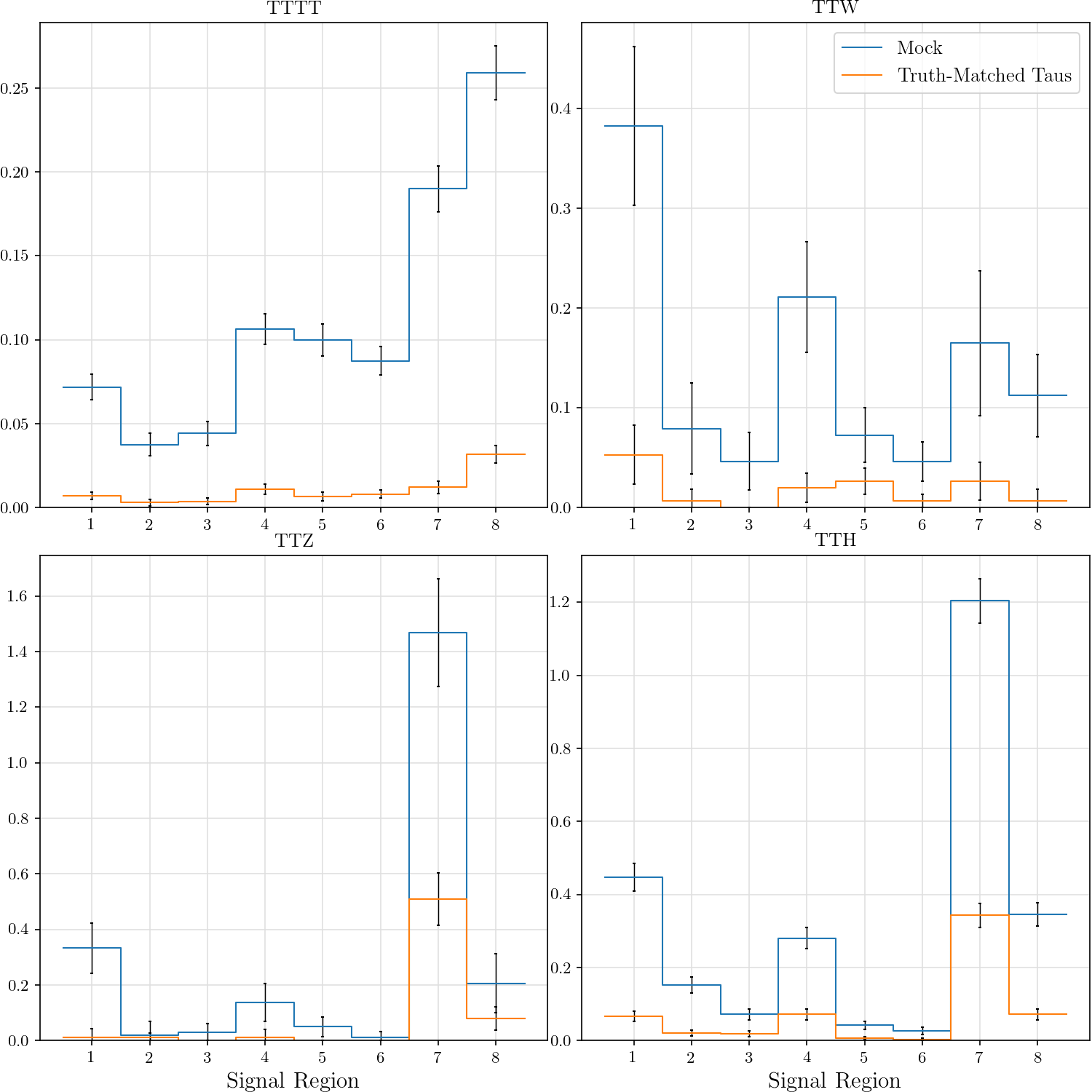
return hists_to_table(hists, row_labels=['TTTT', 'TTW', 'TTZ', 'TTH'],
|
|
|
- column_labels=[f'SR{n}' for n in range(1, 9)])
|
|
|
+ column_labels=[f'SR{n}' for n in range(1, len(tttt[0])+1)])
|
|
|
|
|
|
tables = '<h2>Mock</h2>'
|
|
|
tables += to_table([tttt, ttw, ttz, tth])
|
|
|
@@ -108,6 +108,71 @@ def plot_yield_grid(rss, tau_category=-1):
|
|
|
return tables
|
|
|
|
|
|
|
|
|
+@decl_plot
|
|
|
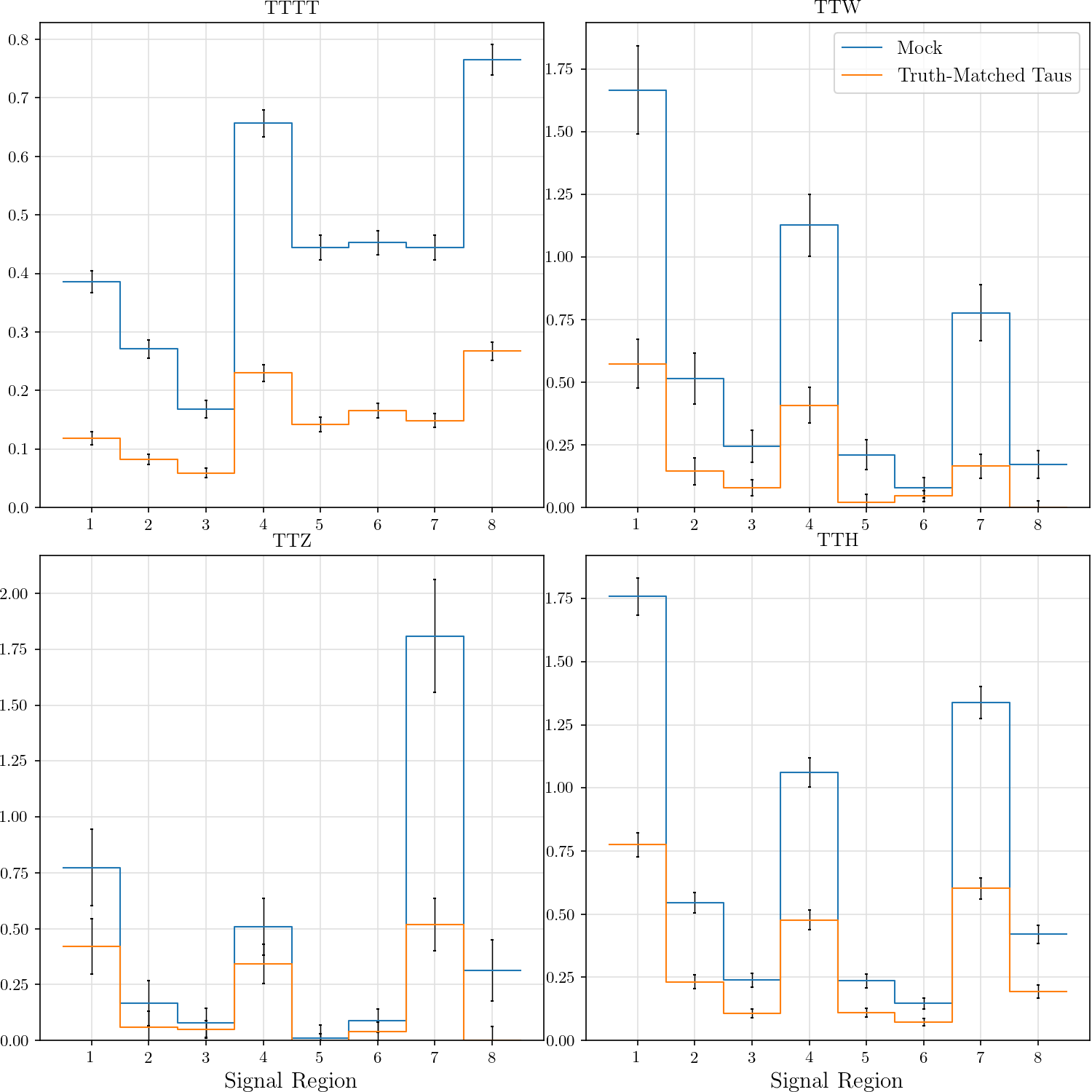
+def plot_yield_v_gen(rss, tau_category=-1):
|
|
|
+ r"""## Event Yield Vs. # of Generated Taus
|
|
|
+
|
|
|
+ """
|
|
|
+ def get_sr(rs):
|
|
|
+ h = None
|
|
|
+ if tau_category == 0:
|
|
|
+ h = rs.SRs_0tmtau_diff_nGenTau
|
|
|
+ if tau_category == 1:
|
|
|
+ h = rs.SRs_1tmtau_diff_nGenTau
|
|
|
+ elif tau_category == 2:
|
|
|
+ h = rs.SRs_2tmtau_diff_nGenTau
|
|
|
+ return hist2d_norm(hist2d(h), norm=1, axis=0)
|
|
|
+
|
|
|
+ _, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
+ tttt, ttw, ttz, tth = [get_sr(rs) for rs in rss]
|
|
|
+
|
|
|
+ def do_plot(h, title):
|
|
|
+ hist2d_plot(h, title=title, txt_format='{:.2f}')
|
|
|
+
|
|
|
+ plt.sca(ax_tttt)
|
|
|
+ do_plot(tttt, 'TTTT')
|
|
|
+ plt.ylabel("\# Gen Taus")
|
|
|
+
|
|
|
+ plt.sca(ax_ttw)
|
|
|
+ do_plot(ttw, 'TTW')
|
|
|
+ plt.legend()
|
|
|
+
|
|
|
+ plt.sca(ax_ttz)
|
|
|
+ do_plot(ttz, 'TTZ')
|
|
|
+ plt.xlabel('Signal Region')
|
|
|
+ plt.ylabel("\# Gen Taus")
|
|
|
+
|
|
|
+ plt.sca(ax_tth)
|
|
|
+ do_plot(tth, 'TTH')
|
|
|
+ plt.xlabel('Signal Region')
|
|
|
+
|
|
|
+
|
|
|
+@decl_plot
|
|
|
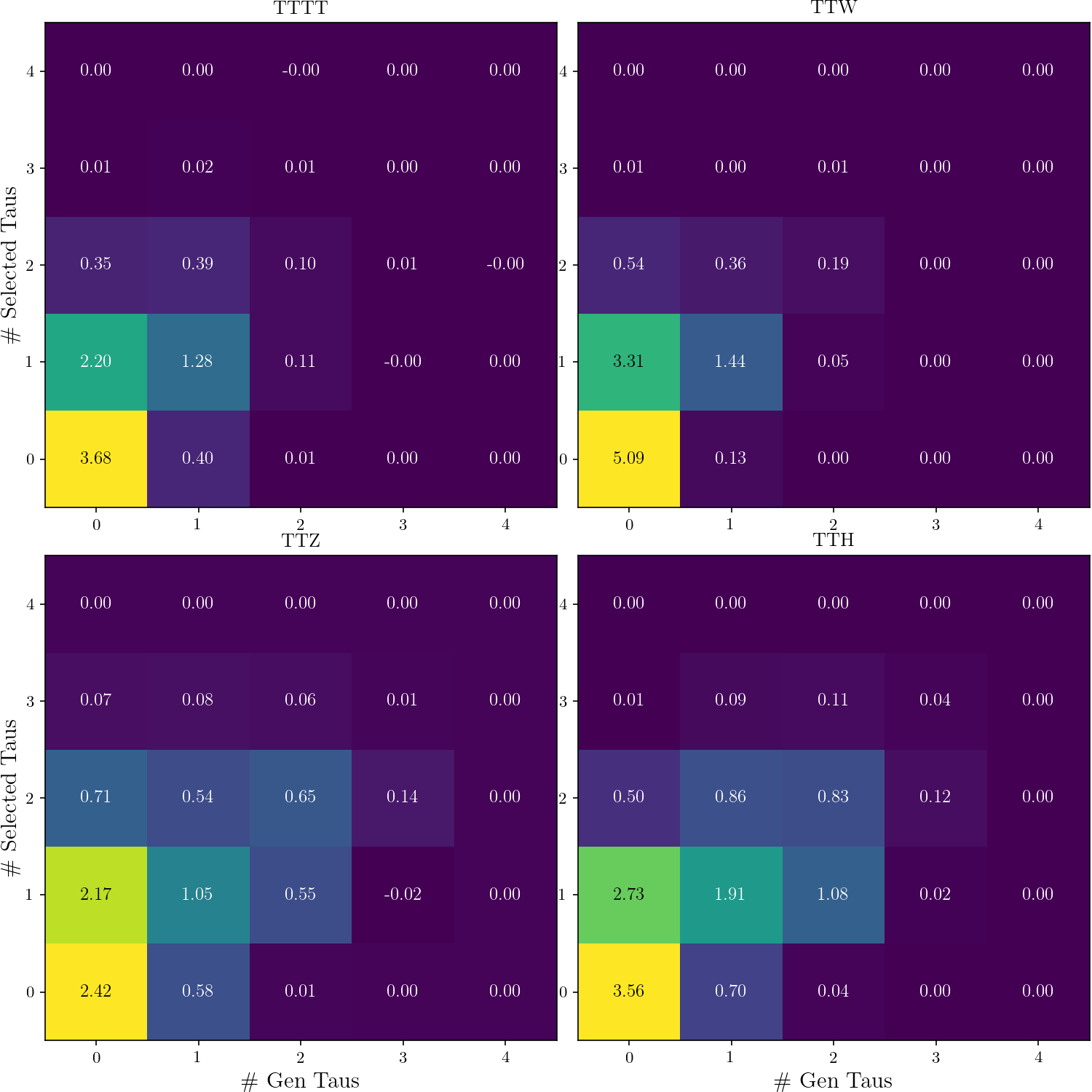
+def plot_nGen_v_nSel(rss):
|
|
|
+ _, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
+ tttt, ttw, ttz, tth = [hist2d(rs.nGen_v_RecoTaus_in_SR) for rs in rss]
|
|
|
+
|
|
|
+ def do_plot(h, title):
|
|
|
+ hist2d_plot(h, title=title, txt_format='{:.2f}')
|
|
|
+
|
|
|
+ plt.sca(ax_tttt)
|
|
|
+ do_plot(tttt, 'TTTT')
|
|
|
+ plt.ylabel("\# Selected Taus")
|
|
|
+
|
|
|
+ plt.sca(ax_ttw)
|
|
|
+ do_plot(ttw, 'TTW')
|
|
|
+ plt.legend()
|
|
|
+
|
|
|
+ plt.sca(ax_ttz)
|
|
|
+ do_plot(ttz, 'TTZ')
|
|
|
+ plt.xlabel('\# Gen Taus')
|
|
|
+ plt.ylabel("\# Selected Taus")
|
|
|
+
|
|
|
+ plt.sca(ax_tth)
|
|
|
+ do_plot(tth, 'TTH')
|
|
|
+ plt.xlabel('\# Gen Taus')
|
|
|
+
|
|
|
+
|
|
|
@decl_plot
|
|
|
def plot_yield_stack(rss):
|
|
|
r"""## Event Yield - Stacked
|
|
|
@@ -116,10 +181,10 @@ def plot_yield_stack(rss):
|
|
|
to the Moriond 2018 integrated luminosity ($35.9\textrm{fb}^{-1}$). Code for the histogram generation is
|
|
|
here: <https://github.com/cfangmeier/FTAnalysis/blob/master/studies/tau/Yield.C>
|
|
|
"""
|
|
|
- ft, ttw, ttz, tth = map(lambda rs: hist(rs.SRs), rss)
|
|
|
+ tttt, ttw, ttz, tth = map(lambda rs: hist(rs.SRs), rss)
|
|
|
hist_plot_stack([ttw, ttz, tth], labels=['TTW', 'TTZ', 'TTH'])
|
|
|
- ft = ft[0]*10, ft[1], ft[2]
|
|
|
- hist_plot(ft, label='TTTT (x10)', stats=False, color='k')
|
|
|
+ tttt = tttt[0]*10, tttt[1], tttt[2]
|
|
|
+ hist_plot(tttt, label='TTTT (x10)', stats=False, color='k')
|
|
|
|
|
|
plt.ylim((0, 60))
|
|
|
plt.xlabel('Signal Region')
|
|
|
@@ -129,14 +194,14 @@ def plot_yield_stack(rss):
|
|
|
@decl_plot
|
|
|
def plot_lep_multi(rss, dataset):
|
|
|
_, (ax_els, ax_mus, ax_taus) = plt.subplots(3, 1)
|
|
|
- els = list(map(lambda rs: hist_normalize(hist(rs.nEls)), rss))
|
|
|
- mus = list(map(lambda rs: hist_normalize(hist(rs.nMus)), rss))
|
|
|
- taus = list(map(lambda rs: hist_normalize(hist(rs.nTaus)), rss))
|
|
|
+ els = list(map(lambda rs: hist_norm(hist(rs.nEls)), rss))
|
|
|
+ mus = list(map(lambda rs: hist_norm(hist(rs.nMus)), rss))
|
|
|
+ taus = list(map(lambda rs: hist_norm(hist(rs.nTaus)), rss))
|
|
|
|
|
|
def _plot(ax, procs):
|
|
|
plt.sca(ax)
|
|
|
- ft, ttw, ttz, tth = procs
|
|
|
- h = {'TTTT': ft,
|
|
|
+ tttt, ttw, ttz, tth = procs
|
|
|
+ h = {'TTTT': tttt,
|
|
|
'TTW': ttw,
|
|
|
'TTZ': ttz,
|
|
|
'TTH': tth}[dataset]
|
|
|
@@ -158,10 +223,10 @@ def plot_sig_strength(rss):
|
|
|
$\frac{S}{\sqrt{S+B}}$
|
|
|
|
|
|
"""
|
|
|
- ft, ttw, ttz, tth = map(lambda rs: hist(rs.SRs), rss)
|
|
|
+ tttt, ttw, ttz, tth = map(lambda rs: hist(rs.SRs), rss)
|
|
|
bg = hist_add(ttw, ttz, tth)
|
|
|
- strength = ft[0] / np.sqrt(ft[0] + bg[0])
|
|
|
- hist_plot((strength, ft[1], ft[2]), stats=False)
|
|
|
+ strength = tttt[0] / np.sqrt(tttt[0] + bg[0])
|
|
|
+ hist_plot((strength, tttt[1], tttt[2]), stats=False)
|
|
|
|
|
|
|
|
|
@decl_plot
|
|
|
@@ -171,9 +236,9 @@ def plot_event_obs(rss, dataset, in_signal_region=True):
|
|
|
|
|
|
"""
|
|
|
_, ((ax_njet, ax_nbjet), (ax_ht, ax_met)) = plt.subplots(2, 2)
|
|
|
- # ft, ttw, ttz, tth = map(lambda rs: hist(rs.SRs), rss)
|
|
|
- ft, ttw, ttz, tth = rss
|
|
|
- rs = {'TTTT': ft,
|
|
|
+ # tttt, ttw, ttz, tth = map(lambda rs: hist(rs.SRs), rss)
|
|
|
+ tttt, ttw, ttz, tth = rss
|
|
|
+ rs = {'TTTT': tttt,
|
|
|
'TTW': ttw,
|
|
|
'TTZ': ttz,
|
|
|
'TTH': tth}[dataset]
|
|
|
@@ -217,9 +282,9 @@ def plot_event_obs_stack(rss, in_signal_region=True):
|
|
|
'HT': 'ht',
|
|
|
'NJET': 'njet',
|
|
|
'NBJET': 'nbjet'}[obs]
|
|
|
- ft, ttw, ttz, tth = map(lambda rs: hist(getattr(rs, attr)), rss)
|
|
|
+ tttt, ttw, ttz, tth = map(lambda rs: hist(getattr(rs, attr)), rss)
|
|
|
hist_plot_stack([ttw, ttz, tth], labels=["TTW", "TTZ", "TTH"])
|
|
|
- hist_plot(hist_scale(ft, 5), label="TTTT (x5)", color='k')
|
|
|
+ hist_plot(hist_scale(tttt, 5), label="TTTT (x5)", color='k')
|
|
|
plt.xlabel(obs)
|
|
|
|
|
|
_plot(ax_njet, 'NJET')
|
|
|
@@ -228,15 +293,31 @@ def plot_event_obs_stack(rss, in_signal_region=True):
|
|
|
_plot(ax_ht, 'HT')
|
|
|
_plot(ax_met, 'MET')
|
|
|
|
|
|
+# @decl_plot
|
|
|
+# def plot_s_over_b(rss):
|
|
|
+# def get_sr(rs):
|
|
|
+# h = None
|
|
|
+# if tau_category == 0:
|
|
|
+# h = rs.SRs_0tmtau_diff_nGenTau
|
|
|
+# if tau_category == 1:
|
|
|
+# h = rs.SRs_1tmtau_diff_nGenTau
|
|
|
+# elif tau_category == 2:
|
|
|
+# h = rs.SRs_2tmtau_diff_nGenTau
|
|
|
+# return hist2d_norm(hist2d(h), norm=1, axis=0)
|
|
|
+#
|
|
|
+# _, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
+# tttt =
|
|
|
+# ttw, ttz, tth = [get_sr(rs) for rs in rss]
|
|
|
+# pass
|
|
|
|
|
|
@decl_plot
|
|
|
def plot_tau_purity(rss):
|
|
|
- _, ((ax_ft, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
- ft, ttw, ttz, tth = list(map(lambda rs: hist(rs.tau_purity_v_pt), rss))
|
|
|
+ _, ((ax_tttt, ax_ttw), (ax_ttz, ax_tth)) = plt.subplots(2, 2)
|
|
|
+ tttt, ttw, ttz, tth = list(map(lambda rs: hist(rs.tau_purity_v_pt), rss))
|
|
|
|
|
|
def _plot(ax, dataset):
|
|
|
plt.sca(ax)
|
|
|
- h = {'TTTT': ft,
|
|
|
+ h = {'TTTT': tttt,
|
|
|
'TTW': ttw,
|
|
|
'TTZ': ttz,
|
|
|
'TTH': tth}[dataset]
|
|
|
@@ -244,19 +325,21 @@ def plot_tau_purity(rss):
|
|
|
plt.text(200, 0.05, dataset)
|
|
|
plt.xlabel(r"$P_T$(GeV)")
|
|
|
|
|
|
- _plot(ax_ft, 'TTTT')
|
|
|
+ _plot(ax_tttt, 'TTTT')
|
|
|
_plot(ax_ttw, 'TTW')
|
|
|
_plot(ax_ttz, 'TTZ')
|
|
|
_plot(ax_tth, 'TTH')
|
|
|
|
|
|
|
|
|
if __name__ == '__main__':
|
|
|
+ data_path = 'data/output_new_sr_new_id_binning/'
|
|
|
+ save_plots = True
|
|
|
# First create a ResultSet object which loads all of the objects from root file
|
|
|
# into memory and makes them available as attributes
|
|
|
- rss = (ResultSet("ft", 'data/yield_ft.root'),
|
|
|
- ResultSet("ttw", 'data/yield_ttw.root'),
|
|
|
- ResultSet("ttz", 'data/yield_ttz.root'),
|
|
|
- ResultSet("tth", 'data/yield_tth.root'))
|
|
|
+ rss = (ResultSet("tttt", data_path+'yield_tttt.root'),
|
|
|
+ ResultSet("ttw", data_path+'yield_ttw.root'),
|
|
|
+ ResultSet("ttz", data_path+'yield_ttz.root'),
|
|
|
+ ResultSet("tth", data_path+'yield_tth.root'))
|
|
|
|
|
|
# Next, declare all of the (sub)plots that will be assembled into full
|
|
|
# figures later
|
|
|
@@ -265,22 +348,26 @@ if __name__ == '__main__':
|
|
|
yield_tau_1tau = plot_yield_grid, (rss, 1)
|
|
|
yield_tau_2tau = plot_yield_grid, (rss, 2)
|
|
|
|
|
|
- ft_event_obs_in_sr = plot_event_obs, (rss, 'TTTT'), {'in_signal_region': True}
|
|
|
+ tttt_event_obs_in_sr = plot_event_obs, (rss, 'TTTT'), {'in_signal_region': True}
|
|
|
ttw_event_obs_in_sr = plot_event_obs, (rss, 'TTW'), {'in_signal_region': True}
|
|
|
ttz_event_obs_in_sr = plot_event_obs, (rss, 'TTZ'), {'in_signal_region': True}
|
|
|
tth_event_obs_in_sr = plot_event_obs, (rss, 'TTH'), {'in_signal_region': True}
|
|
|
|
|
|
- ft_event_obs = plot_event_obs, (rss, 'TTTT'), {'in_signal_region': False}
|
|
|
+ tttt_event_obs = plot_event_obs, (rss, 'TTTT'), {'in_signal_region': False}
|
|
|
ttw_event_obs = plot_event_obs, (rss, 'TTW'), {'in_signal_region': False}
|
|
|
ttz_event_obs = plot_event_obs, (rss, 'TTZ'), {'in_signal_region': False}
|
|
|
tth_event_obs = plot_event_obs, (rss, 'TTH'), {'in_signal_region': False}
|
|
|
|
|
|
- ft_lep_multi = plot_lep_multi, (rss, 'TTTT')
|
|
|
+ tttt_lep_multi = plot_lep_multi, (rss, 'TTTT')
|
|
|
ttw_lep_multi = plot_lep_multi, (rss, 'TTW')
|
|
|
ttz_lep_multi = plot_lep_multi, (rss, 'TTZ')
|
|
|
tth_lep_multi = plot_lep_multi, (rss, 'TTH')
|
|
|
|
|
|
+ yield_v_gen_0tm = plot_yield_v_gen, (rss, 0)
|
|
|
+ yield_v_gen_1tm = plot_yield_v_gen, (rss, 1)
|
|
|
+ yield_v_gen_2tm = plot_yield_v_gen, (rss, 2)
|
|
|
# tau_purity = plot_tau_purity, (rss)
|
|
|
+ nGen_v_nSel = plot_nGen_v_nSel, (rss,)
|
|
|
|
|
|
# Now assemble the plots into figures.
|
|
|
plots = [
|
|
|
@@ -293,7 +380,16 @@ if __name__ == '__main__':
|
|
|
Plot([[yield_tau_2tau]],
|
|
|
'Yield For events with 2 or more Tau'),
|
|
|
|
|
|
- Plot([[ft_lep_multi]],
|
|
|
+ # Plot([[yield_v_gen_0tm]],
|
|
|
+ # 'Yield For events with 0 TM Tau Vs Gen Tau'),
|
|
|
+ # Plot([[yield_v_gen_1tm]],
|
|
|
+ # 'Yield For events with 1 TM Tau Vs Gen Tau'),
|
|
|
+ # Plot([[yield_v_gen_2tm]],
|
|
|
+ # 'Yield For events with 2 TM Tau Vs Gen Tau'),
|
|
|
+ Plot([[nGen_v_nSel]],
|
|
|
+ r'#Generated tau vs. #Selected tau'),
|
|
|
+
|
|
|
+ Plot([[tttt_lep_multi]],
|
|
|
'Lepton Multiplicity - TTTT'),
|
|
|
Plot([[ttw_lep_multi]],
|
|
|
'Lepton Multiplicity - TTW'),
|
|
|
@@ -301,7 +397,7 @@ if __name__ == '__main__':
|
|
|
'Lepton Multiplicity - TTZ'),
|
|
|
Plot([[tth_lep_multi]],
|
|
|
'Lepton Multiplicity - TTH'),
|
|
|
- Plot([[ft_event_obs_in_sr]],
|
|
|
+ Plot([[tttt_event_obs_in_sr]],
|
|
|
'TTTT - Event Observables (In SR)'),
|
|
|
Plot([[ttw_event_obs_in_sr]],
|
|
|
'TTW - Event Observables (In SR)'),
|
|
|
@@ -309,14 +405,14 @@ if __name__ == '__main__':
|
|
|
'TTZ - Event Observables (In SR)'),
|
|
|
Plot([[tth_event_obs_in_sr]],
|
|
|
'TTH - Event Observables (In SR)'),
|
|
|
- Plot([[ft_event_obs]],
|
|
|
- 'TTTT - Event Observables (All Events)'),
|
|
|
- Plot([[ttw_event_obs]],
|
|
|
- 'TTW - Event Observables (All Events)'),
|
|
|
- Plot([[ttz_event_obs]],
|
|
|
- 'TTZ - Event Observables (All Events)'),
|
|
|
- Plot([[tth_event_obs]],
|
|
|
- 'TTH - Event Observables (All Events)'),
|
|
|
+ # Plot([[tttt_event_obs]],
|
|
|
+ # 'TTTT - Event Observables (All Events)'),
|
|
|
+ # Plot([[ttw_event_obs]],
|
|
|
+ # 'TTW - Event Observables (All Events)'),
|
|
|
+ # Plot([[ttz_event_obs]],
|
|
|
+ # 'TTZ - Event Observables (All Events)'),
|
|
|
+ # Plot([[tth_event_obs]],
|
|
|
+ # 'TTH - Event Observables (All Events)'),
|
|
|
|
|
|
# Plot([[yield_notau]],
|
|
|
# 'Yield Without Tau'),
|
|
|
@@ -338,6 +434,12 @@ if __name__ == '__main__':
|
|
|
|
|
|
# Finally, render and save the plots and generate the html+bootstrap
|
|
|
# dashboard to view them
|
|
|
- render_plots(plots, to_disk=False)
|
|
|
- generate_dashboard(plots, 'TTTT Yields', output='yield_breakout_4.html', source_file=__file__,
|
|
|
- ana_source="https://github.com/cfangmeier/FTAnalysis/commit/46fc1afab42f6b04c9e21ba519e6a30524983550")
|
|
|
+ render_plots(plots, to_disk=save_plots)
|
|
|
+ if not save_plots:
|
|
|
+ generate_dashboard(plots, 'TTTT Yields',
|
|
|
+ output='yields.html',
|
|
|
+ source=__file__,
|
|
|
+ ana_source=("https://github.com/cfangmeier/FTAnalysis/commit/"
|
|
|
+ "0cbdac4509391fffb9fff87d0521b7dd0a30a55c"),
|
|
|
+ config=data_path+'config.yaml'
|
|
|
+ )
|